[English] 日本語
 Yorodumi
Yorodumi- EMDB-4173: A FliPQR complex forms the core of the Salmonella type III secret... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4173 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | A FliPQR complex forms the core of the Salmonella type III secretion system export apparatus. | |||||||||||||||
 Map data Map data | Post-processed (B-factor sharpened and masked) map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | T3SS / Flagella / Cryo-EM / Export / Membrane protein / PROTEIN TRANSPORT | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationbacterial-type flagellum organization / bacterial-type flagellum basal body / bacterial-type flagellum-dependent swarming motility / bacterial-type flagellum assembly / protein secretion / protein targeting / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Salmonella enterica (bacteria) / Salmonella enterica (bacteria) /  Salmonella enterica subsp. enterica (bacteria) / Salmonella enterica subsp. enterica (bacteria) /  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) | |||||||||||||||
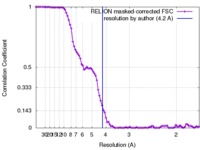
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||||||||
 Authors Authors | Johnson S / Kuhlen L | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Structure of the core of the type III secretion system export apparatus. Authors: Lucas Kuhlen / Patrizia Abrusci / Steven Johnson / Joseph Gault / Justin Deme / Joseph Caesar / Tobias Dietsche / Mehari Tesfazgi Mebrhatu / Tariq Ganief / Boris Macek / Samuel Wagner / ...Authors: Lucas Kuhlen / Patrizia Abrusci / Steven Johnson / Joseph Gault / Justin Deme / Joseph Caesar / Tobias Dietsche / Mehari Tesfazgi Mebrhatu / Tariq Ganief / Boris Macek / Samuel Wagner / Carol V Robinson / Susan M Lea /   Abstract: Export of proteins through type III secretion systems is critical for motility and virulence of many major bacterial pathogens. Three putative integral membrane proteins (FliP, FliQ, FliR) are ...Export of proteins through type III secretion systems is critical for motility and virulence of many major bacterial pathogens. Three putative integral membrane proteins (FliP, FliQ, FliR) are suggested to form the core of an export gate in the inner membrane, but their structure, assembly and location within the final nanomachine remain unclear. Here, we present the cryoelectron microscopy structure of the Salmonella Typhimurium FliP-FliQ-FliR complex at 4.2 Å. None of the subunits adopt canonical integral membrane protein topologies, and common helix-turn-helix structural elements allow them to form a helical assembly with 5:4:1 stoichiometry. Fitting of the structure into reconstructions of intact secretion systems, combined with cross-linking, localize the export gate as a core component of the periplasmic portion of the machinery. This study thereby identifies the export gate as a key element of the secretion channel and implies that it primes the helical architecture of the components assembling downstream. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4173.map.gz emd_4173.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4173-v30.xml emd-4173-v30.xml emd-4173.xml emd-4173.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4173_fsc.xml emd_4173_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_4173.png emd_4173.png | 62.9 KB | ||
| Filedesc metadata |  emd-4173.cif.gz emd-4173.cif.gz | 6.8 KB | ||
| Others |  emd_4173_half_map_1.map.gz emd_4173_half_map_1.map.gz emd_4173_half_map_2.map.gz emd_4173_half_map_2.map.gz | 49.4 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4173 http://ftp.pdbj.org/pub/emdb/structures/EMD-4173 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4173 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4173 | HTTPS FTP |
-Related structure data
| Related structure data |  6f2dMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4173.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4173.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed (B-factor sharpened and masked) map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1
| File | emd_4173_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
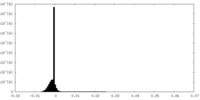
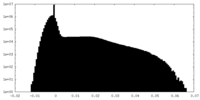
| Density Histograms |
-Half map: Half map 2
| File | emd_4173_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
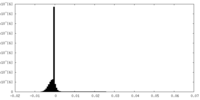
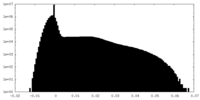
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of the flagellar type III secretion system export apparat...
| Entire | Name: Complex of the flagellar type III secretion system export apparatus components FliP, FliQ and FliR |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the flagellar type III secretion system export apparat...
| Supramolecule | Name: Complex of the flagellar type III secretion system export apparatus components FliP, FliQ and FliR type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Salmonella enterica (bacteria) Salmonella enterica (bacteria) |
| Molecular weight | Theoretical: 195 kDa/nm |
-Macromolecule #1: Flagellar biosynthetic protein FliP
| Macromolecule | Name: Flagellar biosynthetic protein FliP / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica (bacteria) Salmonella enterica subsp. enterica (bacteria) |
| Molecular weight | Theoretical: 26.769021 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRRLLFLSLA GLWLFSPAAA AQLPGLISQP LAGGGQSWSL SVQTLVFITS LTFLPAILLM MTSFTRIIIV FGLLRNALGT PSAPPNQVL LGLALFLTFF IMSPVIDKIY VDAYQPFSEQ KISMQEALDK GAQPLRAFML RQTREADLAL FARLANSGPL Q GPEAVPMR ...String: MRRLLFLSLA GLWLFSPAAA AQLPGLISQP LAGGGQSWSL SVQTLVFITS LTFLPAILLM MTSFTRIIIV FGLLRNALGT PSAPPNQVL LGLALFLTFF IMSPVIDKIY VDAYQPFSEQ KISMQEALDK GAQPLRAFML RQTREADLAL FARLANSGPL Q GPEAVPMR ILLPAYVTSE LKTAFQIGFT IFIPFLIIDL VIASVLMALG MMMVPPATIA LPFKLMLFVL VDGWQLLVGS LA QSFYS UniProtKB: Flagellar biosynthetic protein FliP |
-Macromolecule #2: Flagellar biosynthetic protein FliR
| Macromolecule | Name: Flagellar biosynthetic protein FliR / type: protein_or_peptide / ID: 2 Details: Residues 265 onwards constitute the Twin-Strep tag used for purification Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 33.06918 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIQVTSEQWL YWLHLYFWPL LRVLALISTA PILSERAIPK RVKLGLGIMI TLVIAPSLPA NDTPLFSIAA LWLAMQQILI GIALGFTMQ FAFAAVRTAG EFIGLQMGLS FATFVDPGSH LNMPVLARIM DMLAMLLFLT FNGHLWLISL LVDTFHTLPI G SNPVNSNA ...String: MIQVTSEQWL YWLHLYFWPL LRVLALISTA PILSERAIPK RVKLGLGIMI TLVIAPSLPA NDTPLFSIAA LWLAMQQILI GIALGFTMQ FAFAAVRTAG EFIGLQMGLS FATFVDPGSH LNMPVLARIM DMLAMLLFLT FNGHLWLISL LVDTFHTLPI G SNPVNSNA FMALARAGGL IFLNGLMLAL PVITLLLTLN LALGLLNRMA PQLSIFVIGF PLTLTVGIML MAALMPLIAP FC EHLFSEI FNLLADIVSE MPINNNPENL YFQGQFGSWS HPQFEKGGGS GGGSGGGSWS HPQFEK UniProtKB: Flagellar biosynthetic protein FliR |
-Macromolecule #3: Flagellar biosynthetic protein FliQ
| Macromolecule | Name: Flagellar biosynthetic protein FliQ / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) |
| Molecular weight | Theoretical: 9.606758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTPESVMMMG TEAMKVALAL AAPLLLVALI TGLIISILQA ATQINEMTLS FIPKIVAVFI AIIVAGPWML NLLLDYVRTL FSNLPYIIG UniProtKB: Flagellar biosynthetic protein FliQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Detector mode: COUNTING / #0 - Number real images: 401 / #0 - Average electron dose: 47.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: FEI FALCON III (4k x 4k) / #1 - Detector mode: COUNTING / #1 - Number real images: 1687 / #1 - Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)