+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3784 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the membrane-bound human mitoribosome | ||||||||||||
 Map data Map data | Structure of the membrane-bound human mitoribosome in isolated mitochondria | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 26.0 Å | ||||||||||||
 Authors Authors | Englmeier R / Pfeffer S / Foerster F | ||||||||||||
| Funding support |  Germany, Germany,  Netherlands, 3 items Netherlands, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: Structure of the Human Mitochondrial Ribosome Studied In Situ by Cryoelectron Tomography. Authors: Robert Englmeier / Stefan Pfeffer / Friedrich Förster /   Abstract: Mitochondria maintain their own genome and its corresponding protein synthesis machine, the mitochondrial ribosome (mitoribosome). Mitoribosomes primarily synthesize highly hydrophobic proteins ...Mitochondria maintain their own genome and its corresponding protein synthesis machine, the mitochondrial ribosome (mitoribosome). Mitoribosomes primarily synthesize highly hydrophobic proteins of the inner mitochondrial membrane. Recent studies revealed the complete structure of the isolated mammalian mitoribosome, but its mode of membrane association remained hypothetical. In this study, we used cryoelectron tomography to visualize human mitoribosomes in isolated mitochondria. The subtomogram average of the membrane-associated human mitoribosome reveals a single major contact site with the inner membrane, mediated by the mitochondria-specific protein mL45. A second rRNA-mediated contact site that is present in yeast is absent in humans, resulting in a more variable association of the human mitoribosome with the inner membrane. Despite extensive structural differences of mammalian and fungal mitoribosomal structure, the principal organization of peptide exit tunnel and the mL45 homolog remains invariant, presumably to align the mitoribosome with the membrane-embedded insertion machinery. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3784.map.gz emd_3784.map.gz | 4.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3784-v30.xml emd-3784-v30.xml emd-3784.xml emd-3784.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3784.png emd_3784.png | 1.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3784 http://ftp.pdbj.org/pub/emdb/structures/EMD-3784 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3784 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3784 | HTTPS FTP |
-Validation report
| Summary document |  emd_3784_validation.pdf.gz emd_3784_validation.pdf.gz | 227 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3784_full_validation.pdf.gz emd_3784_full_validation.pdf.gz | 226.2 KB | Display | |
| Data in XML |  emd_3784_validation.xml.gz emd_3784_validation.xml.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3784 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3784 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3784 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3784 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3784.map.gz / Format: CCP4 / Size: 5.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3784.map.gz / Format: CCP4 / Size: 5.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the membrane-bound human mitoribosome in isolated mitochondria | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
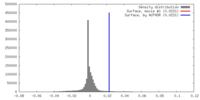
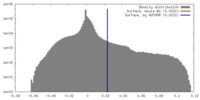
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Whole mitochondria purified from human cell culture
| Entire | Name: Whole mitochondria purified from human cell culture |
|---|---|
| Components |
|
-Supramolecule #1: Whole mitochondria purified from human cell culture
| Supramolecule | Name: Whole mitochondria purified from human cell culture / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: HEK-293 / Organelle: Mitochondria Homo sapiens (human) / Strain: HEK-293 / Organelle: Mitochondria |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 1.2 sec. / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)