+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2826 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the membrane-bound mitoribosome from yeast | |||||||||
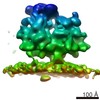
 Map data Map data | Subtomogram average of the membrane-bound mitoribosome from yeast | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / mitoribosome / mitochondria / Mba1 / tomography subtomogram analysis | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / negative staining / Resolution: 30.0 Å | |||||||||
 Authors Authors | Pfeffer S / Woellhaf MW / Herrmann JM / Foerster F | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2015 Journal: Nat Commun / Year: 2015Title: Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Authors: Stefan Pfeffer / Michael W Woellhaf / Johannes M Herrmann / Friedrich Förster /  Abstract: Whereas the structure and function of cytosolic ribosomes have been studied in great detail, we know surprisingly little about the structural basis of mitochondrial protein synthesis. Here we used ...Whereas the structure and function of cytosolic ribosomes have been studied in great detail, we know surprisingly little about the structural basis of mitochondrial protein synthesis. Here we used cryoelectron tomography and subtomogram analysis to visualize mitoribosomes in isolated yeast mitochondria, avoiding perturbations during ribosomal purification. Most mitoribosomes reside in immediate proximity to the inner mitochondrial membrane, in line with their specialization in the synthesis of hydrophobic membrane proteins. The subtomogram average of membrane-associated mitoribosomes reveals two distinct membrane contact sites, formed by the 21S rRNA expansion segment 96-ES1 and the inner membrane protein Mba1. On the basis of our data, we further hypothesize that Mba1 is not just a passive mitoribosome receptor on the inner membrane, but that it spatially aligns mitoribosomes with the membrane insertion machinery. This study reveals detailed insights into the supramolecular organization of the mitochondrial translation machinery and its association with the inner membrane in translation-competent mitochondria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2826.map.gz emd_2826.map.gz | 28.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2826-v30.xml emd-2826-v30.xml emd-2826.xml emd-2826.xml | 8.5 KB 8.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2826.tif emd_2826.tif | 171.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2826 http://ftp.pdbj.org/pub/emdb/structures/EMD-2826 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2826 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2826 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2826.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2826.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of the membrane-bound mitoribosome from yeast | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Membrane-bound mitoribosome in isolated mitochondria from yeast
| Entire | Name: Membrane-bound mitoribosome in isolated mitochondria from yeast |
|---|---|
| Components |
|
-Supramolecule #1000: Membrane-bound mitoribosome in isolated mitochondria from yeast
| Supramolecule | Name: Membrane-bound mitoribosome in isolated mitochondria from yeast type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Membrane-bound 73S mitoribosome
| Supramolecule | Name: Membrane-bound 73S mitoribosome / type: complex / ID: 1 / Name.synonym: 73S mitoribosome / Recombinant expression: No / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 / Details: 20 mM Hepes pH 7.6, 50 mM KCl, 2 mM MgCl2 |
|---|---|
| Staining | Type: NEGATIVE / Details: No staining |
| Grid | Details: Lacey carbon molybdenum grids |
| Vitrification | Cryogen name: ETHANE-PROPANE MIXTURE / Chamber humidity: 60 % / Instrument: FEI VITROBOT MARK IV / Method: Blot time: 4s; blot force: 0 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Nov 10, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Average electron dose: 100 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 7.0 µm / Nominal defocus min: 5.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Tomogram reconstruction and template matching against a single particle cryo-EM reconstruction of the 73S yeast mitoribosome were accomplished using PyTom. Tomogram areas corresponding to cross correlation peaks within mitochondria were visually inspected to identify true positive matches. For the retained coordinates, unbinned subtomograms were reconstructed individually from the weighted projections and iteratively aligned using PyTom. Aligned subtomograms were classified using constrained principal component analysis. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: PyTom, tom_toolbox, av3_toolbox / Number subtomograms used: 900 |
| CTF correction | Details: each micrograph |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















