[English] 日本語
 Yorodumi
Yorodumi- EMDB-6863: 3.9 angstrom cryo-EM structure of human ATR/ATRIP catalytic core -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6863 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3.9 angstrom cryo-EM structure of human ATR/ATRIP catalytic core | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
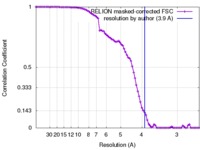
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Rao Q / Liu M / Tian Y / Wu Z / Wang H / Wang J / Xu Y | |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2018 Journal: Cell Res / Year: 2018Title: Cryo-EM structure of human ATR-ATRIP complex. Authors: Qinhui Rao / Mengjie Liu / Yuan Tian / Zihan Wu / Yuhan Hao / Lei Song / Zhaoyu Qin / Chen Ding / Hong-Wei Wang / Jiawei Wang / Yanhui Xu /  Abstract: ATR (ataxia telangiectasia-mutated and Rad3-related) protein kinase and ATRIP (ATR-interacting protein) form a complex and play a critical role in response to replication stress and DNA damage. Here, ...ATR (ataxia telangiectasia-mutated and Rad3-related) protein kinase and ATRIP (ATR-interacting protein) form a complex and play a critical role in response to replication stress and DNA damage. Here, we determined the cryo-electron microscopy (EM) structure of the human ATR-ATRIP complex at 4.7 Å resolution and built an atomic model of the C-terminal catalytic core of ATR (residues 1 521-2 644) at 3.9 Å resolution. The complex adopts a hollow "heart" shape, consisting of two ATR monomers in distinct conformations. The EM map for ATRIP reveals 14 HEAT repeats in an extended "S" shape. The conformational flexibility of ATR allows ATRIP to properly lock the N-termini of the two ATR monomers to favor ATR-ATRIP complex formation and functional diversity. The isolated "head-head" and "tail-tail" each adopts a pseudo 2-fold symmetry. The catalytic pockets face outward and substrate access is not restricted by inhibitory elements. Our studies provide a structural basis for understanding the assembly of the ATR-ATRIP complex and a framework for characterizing ATR-mediated DNA repair pathways. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6863.map.gz emd_6863.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6863-v30.xml emd-6863-v30.xml emd-6863.xml emd-6863.xml | 8.6 KB 8.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6863_fsc.xml emd_6863_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_6863.png emd_6863.png | 73 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6863 http://ftp.pdbj.org/pub/emdb/structures/EMD-6863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6863 | HTTPS FTP |
-Validation report
| Summary document |  emd_6863_validation.pdf.gz emd_6863_validation.pdf.gz | 79.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6863_full_validation.pdf.gz emd_6863_full_validation.pdf.gz | 78.5 KB | Display | |
| Data in XML |  emd_6863_validation.xml.gz emd_6863_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6863 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6863 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6863 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6863 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6863.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6863.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human ATR/ATRIP catalytic core
| Entire | Name: human ATR/ATRIP catalytic core |
|---|---|
| Components |
|
-Supramolecule #1: human ATR/ATRIP catalytic core
| Supramolecule | Name: human ATR/ATRIP catalytic core / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Experimental: 700 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 1.562 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)