[English] 日本語
 Yorodumi
Yorodumi- EMDB-3641: Unraveling the self-assembly of Pseudomonas aeruginosa XcpQ secre... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3641 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Unraveling the self-assembly of Pseudomonas aeruginosa XcpQ secretin periplasmic domain provides new molecular insights into T2SS secreton architecture & dynamics | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein secretion by the type II secretion system / type II protein secretion system complex / protein secretion / cell outer membrane / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
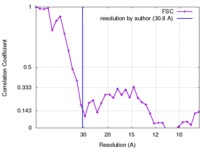
| Method | single particle reconstruction / negative staining / Resolution: 30.8 Å | |||||||||
 Authors Authors | Douzi B / Trinh N / Ball G / Desmyter A / Michel-Souzy S / Barbier P / Kosta A / Durand E / Cambillau C / Roussel A / Voulhoux R | |||||||||
 Citation Citation |  Journal: mBio / Year: 2017 Journal: mBio / Year: 2017Title: Unraveling the Self-Assembly of the XcpQ Secretin Periplasmic Domain Provides New Molecular Insights into Type II Secretion System Secreton Architecture and Dynamics. Authors: Badreddine Douzi / Nhung T T Trinh / Sandra Michel-Souzy / Aline Desmyter / Geneviève Ball / Pascale Barbier / Artemis Kosta / Eric Durand / Katrina T Forest / Christian Cambillau / Alain ...Authors: Badreddine Douzi / Nhung T T Trinh / Sandra Michel-Souzy / Aline Desmyter / Geneviève Ball / Pascale Barbier / Artemis Kosta / Eric Durand / Katrina T Forest / Christian Cambillau / Alain Roussel / Romé Voulhoux /   Abstract: The type II secretion system (T2SS) releases large folded exoproteins across the envelope of many Gram-negative pathogens. This secretion process therefore requires specific gating, interacting, and ...The type II secretion system (T2SS) releases large folded exoproteins across the envelope of many Gram-negative pathogens. This secretion process therefore requires specific gating, interacting, and dynamics properties mainly operated by a bipartite outer membrane channel called secretin. We have a good understanding of the structure-function relationship of the pore-forming C-terminal domain of secretins. In contrast, the high flexibility of their periplasmic N-terminal domain has been an obstacle in obtaining the detailed structural information required to uncover its molecular function. In , the Xcp T2SS plays an important role in bacterial virulence by its capacity to deliver a large panel of toxins and degradative enzymes into the surrounding environment. Here, we revealed that the N-terminal domain of XcpQ secretin spontaneously self-assembled into a hexamer of dimers independently of its C-terminal domain. Furthermore, and by using multidisciplinary approaches, we elucidate the structural organization of the XcpQ N domain and demonstrate that secretin flexibility at interdimer interfaces is mandatory for its function. Bacterial secretins are large homooligomeric proteins constituting the outer membrane pore-forming element of several envelope-embedded nanomachines essential in bacterial survival and pathogenicity. They comprise a well-defined membrane-embedded C-terminal domain and a modular periplasmic N-terminal domain involved in substrate recruitment and connection with inner membrane components. We are studying the XcpQ secretin of the T2SS present in the pathogenic bacterium Our data highlight the ability of the XcpQ N-terminal domain to spontaneously oligomerize into a hexamer of dimers. Further experiments revealed that this domain adopts different conformations essential for the T2SS secretion process. These findings provide new insights into the functional understanding of bacterial T2SS secretins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3641.map.gz emd_3641.map.gz | 374.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3641-v30.xml emd-3641-v30.xml emd-3641.xml emd-3641.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3641_fsc.xml emd_3641_fsc.xml | 3.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_3641_1.png emd_3641_1.png emd_3641_2.png emd_3641_2.png emd_3641_3.png emd_3641_3.png | 18.2 KB 26.7 KB 24.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3641 http://ftp.pdbj.org/pub/emdb/structures/EMD-3641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3641 | HTTPS FTP |
-Validation report
| Summary document |  emd_3641_validation.pdf.gz emd_3641_validation.pdf.gz | 221.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3641_full_validation.pdf.gz emd_3641_full_validation.pdf.gz | 220.3 KB | Display | |
| Data in XML |  emd_3641_validation.xml.gz emd_3641_validation.xml.gz | 6.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3641 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3641 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3641.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3641.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
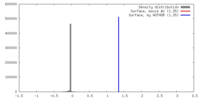
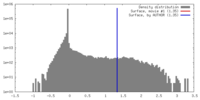
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dodecameric complex of the N-terminal domain of the secretin XcpQ...
| Entire | Name: Dodecameric complex of the N-terminal domain of the secretin XcpQ from Pseudomonas aeruginosa |
|---|---|
| Components |
|
-Supramolecule #1: Dodecameric complex of the N-terminal domain of the secretin XcpQ...
| Supramolecule | Name: Dodecameric complex of the N-terminal domain of the secretin XcpQ from Pseudomonas aeruginosa type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 322 KDa |
-Macromolecule #1: Secretin N terminal domain
| Macromolecule | Name: Secretin N terminal domain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: E N S G G N A F V P A G N Q Q E A H W T I N L K D A D I R E F I D Q I S E I T G E T F V V D P R V K G Q V S V V S K A Q L S L S E V Y Q L F L S V M S T H G F T V V A Q G D Q ...String: E N S G G N A F V P A G N Q Q E A H W T I N L K D A D I R E F I D Q I S E I T G E T F V V D P R V K G Q V S V V S K A Q L S L S E V Y Q L F L S V M S T H G F T V V A Q G D Q A R I V P N A E A K T E A G G G Q S A P D R L E T R V I Q V Q Q S P V S E L I P L I R P L V P Q Y G H L A A V P S A N A L I I S D R S A N I A R I E D V I R Q L D Q K G S H D Y S V I N L R Y G W V M D A A E V L N N A M S R G Q A K G A A G A Q V I A D A R T N R L I I L G P P Q A R A K L V Q L A Q S L D T P |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 / Component:
| ||||||
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: The protein was coated on carbon grid for 3 min. The grid were then washed using drop method 3 times and then incubated with Uranyl Acetate 2% for 3 min. | ||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (2k x 2k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)