+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Synaptic vesicle / epilepsy / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of gamma-aminobutyric acid secretion / Toxicity of botulinum toxin type F (botF) / Toxicity of botulinum toxin type D (botD) / Toxicity of botulinum toxin type E (botE) / Toxicity of botulinum toxin type A (botA) / synaptic vesicle priming / presynaptic active zone / transmembrane transporter activity / protein transmembrane transporter activity / GABA-ergic synapse ...regulation of gamma-aminobutyric acid secretion / Toxicity of botulinum toxin type F (botF) / Toxicity of botulinum toxin type D (botD) / Toxicity of botulinum toxin type E (botE) / Toxicity of botulinum toxin type A (botA) / synaptic vesicle priming / presynaptic active zone / transmembrane transporter activity / protein transmembrane transporter activity / GABA-ergic synapse / neuromuscular junction / metalloendopeptidase activity / synaptic vesicle membrane / intracellular calcium ion homeostasis / cell-cell junction / synaptic vesicle / toxin activity / neuron projection / neuronal cell body / glutamatergic synapse / dendrite / protein kinase binding / endoplasmic reticulum / proteolysis / zinc ion binding / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
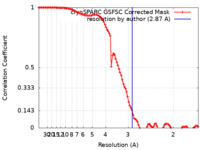
| Method | single particle reconstruction / cryo EM / Resolution: 2.87 Å | |||||||||
 Authors Authors | Yamagata A | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A. Authors: Atsushi Yamagata / Kaori Ito / Takehiro Suzuki / Naoshi Dohmae / Tohru Terada / Mikako Shirouzu /  Abstract: More than one percent of people have epilepsy worldwide. Levetiracetam (LEV) is a successful new-generation antiepileptic drug (AED), and its derivative, brivaracetam (BRV), shows improved efficacy. ...More than one percent of people have epilepsy worldwide. Levetiracetam (LEV) is a successful new-generation antiepileptic drug (AED), and its derivative, brivaracetam (BRV), shows improved efficacy. Synaptic vesicle glycoprotein 2a (SV2A), a putative membrane transporter in the synaptic vesicles (SVs), has been identified as a target of LEV and BRV. SV2A also serves as a receptor for botulinum neurotoxin (BoNT), which is the most toxic protein and has paradoxically emerged as a potent reagent for therapeutic and cosmetic applications. Nevertheless, no structural analysis on AEDs and BoNT recognition by full-length SV2A has been available. Here we describe the cryo-electron microscopy structures of the full-length SV2A in complex with the BoNT receptor-binding domain, BoNT/A2 H and either LEV or BRV. The large fourth luminal domain of SV2A binds to BoNT/A2 H through protein-protein and protein-glycan interactions. LEV and BRV occupy the putative substrate-binding site in an outward-open conformation. A propyl group in BRV creates additional contacts with SV2A, explaining its higher binding affinity than that of LEV, which was further supported by label-free spectral shift assay. Numerous LEV derivatives have been developed as AEDs and positron emission tomography (PET) tracers for neuroimaging. Our work provides a structural framework for AEDs and BoNT recognition of SV2A and a blueprint for the rational design of additional AEDs and PET tracers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36396.map.gz emd_36396.map.gz | 97 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36396-v30.xml emd-36396-v30.xml emd-36396.xml emd-36396.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36396_fsc.xml emd_36396_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36396.png emd_36396.png | 33.8 KB | ||
| Masks |  emd_36396_msk_1.map emd_36396_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36396.cif.gz emd-36396.cif.gz | 6 KB | ||
| Others |  emd_36396_half_map_1.map.gz emd_36396_half_map_1.map.gz emd_36396_half_map_2.map.gz emd_36396_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36396 http://ftp.pdbj.org/pub/emdb/structures/EMD-36396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36396 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36396 | HTTPS FTP |
-Validation report
| Summary document |  emd_36396_validation.pdf.gz emd_36396_validation.pdf.gz | 971 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36396_full_validation.pdf.gz emd_36396_full_validation.pdf.gz | 970.6 KB | Display | |
| Data in XML |  emd_36396_validation.xml.gz emd_36396_validation.xml.gz | 17.8 KB | Display | |
| Data in CIF |  emd_36396_validation.cif.gz emd_36396_validation.cif.gz | 22.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36396 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36396 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36396 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36396 | HTTPS FTP |
-Related structure data
| Related structure data |  8jlgMC  8jlcC  8jleC  8jlfC  8jlhC  8jliC  8js8C  8js9C  8k77C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36396.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36396.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36396_msk_1.map emd_36396_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
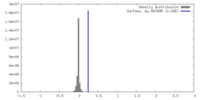
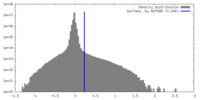
| Density Histograms |
-Half map: #1
| File | emd_36396_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
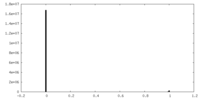
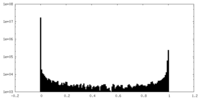
| Density Histograms |
-Half map: #2
| File | emd_36396_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SV2A LD4 in complex with BoNT/A2 Hc domain from the SV2A-BoNT/A2 ...
| Entire | Name: SV2A LD4 in complex with BoNT/A2 Hc domain from the SV2A-BoNT/A2 Hc complex |
|---|---|
| Components |
|
-Supramolecule #1: SV2A LD4 in complex with BoNT/A2 Hc domain from the SV2A-BoNT/A2 ...
| Supramolecule | Name: SV2A LD4 in complex with BoNT/A2 Hc domain from the SV2A-BoNT/A2 Hc complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Synaptic vesicle glycoprotein 2A
| Macromolecule | Name: Synaptic vesicle glycoprotein 2A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.056498 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: YASRTKVFPG ERVEHVTFNF TLENQIHRGG QYFNDKFIGL RLKSVSFEDS LFEECYFEDV TSSNTFFRNC TFINTVFYNT DLFEYKFVN SRLINSTFLH NKEGCPLDVT GT UniProtKB: Synaptic vesicle glycoprotein 2A |
-Macromolecule #2: Botulinum neurotoxin
| Macromolecule | Name: Botulinum neurotoxin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.456191 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KNIVNTSILS IVYKKDDLID LSRYGAKINI GDRVYYDSID KNQIKLINLE SSTIEVILKN AIVYNSMYEN FSTSFWIKIP KYFSKINLN NEYTIINCIE NNSGWKVSLN YGEIIWTLQD NKQNIQRVVF KYSQMVNISD YINRWIFVTI TNNRLTKSKI Y INGRLIDQ ...String: KNIVNTSILS IVYKKDDLID LSRYGAKINI GDRVYYDSID KNQIKLINLE SSTIEVILKN AIVYNSMYEN FSTSFWIKIP KYFSKINLN NEYTIINCIE NNSGWKVSLN YGEIIWTLQD NKQNIQRVVF KYSQMVNISD YINRWIFVTI TNNRLTKSKI Y INGRLIDQ KPISNLGNIH ASNKIMFKLD GCRDPRRYIM IKYFNLFDKE LNEKEIKDLY DSQSNSGILK DFWGNYLQYD KP YYMLNLF DPNKYVDVNN IGIRGYMYLK GPRGSVVTTN IYLNSTLYEG TKFIIKKYAS GNEDNIVRNN DRVYINVVVK NKE YRLATN ASQAGVEKIL SALEIPDVGN LSQVVVMKSK DDQGIRNKCK MNLQDNNGND IGFIGFHLYD NIAKLVASNW YNRQ VGKAS RTFGCSWEFI PVDDGWGESS L UniProtKB: Botulinum neurotoxin |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 20 mM Hepes (pH7.5), 150 mM NaCl, 0.001% LMNG, 0.0002% cholesterol hemisuccinate |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4890 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)