[English] 日本語
 Yorodumi
Yorodumi- EMDB-32900: Cryo-EM structure of Na+,K+-ATPase in the E2P state formed by ino... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Na+,K+-ATPase in the E2P state formed by inorganic phosphate with ouabain | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of monoatomic ion transport / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex / sodium ion export across plasma membrane / intracellular potassium ion homeostasis / intracellular sodium ion homeostasis / potassium ion import across plasma membrane / ATPase activator activity / sodium channel regulator activity / monoatomic ion transport ...regulation of monoatomic ion transport / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex / sodium ion export across plasma membrane / intracellular potassium ion homeostasis / intracellular sodium ion homeostasis / potassium ion import across plasma membrane / ATPase activator activity / sodium channel regulator activity / monoatomic ion transport / proton transmembrane transport / ATP hydrolysis activity / ATP binding / membrane / metal ion binding / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Squalus acanthias (spiny dogfish) / Squalus acanthias (spiny dogfish) /  spiny dogfish (spiny dogfish) spiny dogfish (spiny dogfish) | |||||||||||||||||||||
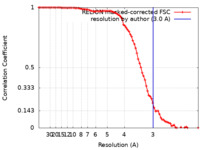
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||||||||
 Authors Authors | Kanai R / Cornelius F / Vilsen B / Toyoshima C | |||||||||||||||||||||
| Funding support |  Japan, Japan,  Denmark, 6 items Denmark, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Cryoelectron microscopy of Na,K-ATPase in the two E2P states with and without cardiotonic steroids. Authors: Ryuta Kanai / Flemming Cornelius / Bente Vilsen / Chikashi Toyoshima /   Abstract: Cryoelectron microscopy (cryo-EM) was applied to Na+,K+-ATPase (NKA) to determine the structures of two E2P states, one (E2PATP) formed by ATP and Mg2+ in the forward reaction, and the other (E2PPi) ...Cryoelectron microscopy (cryo-EM) was applied to Na+,K+-ATPase (NKA) to determine the structures of two E2P states, one (E2PATP) formed by ATP and Mg2+ in the forward reaction, and the other (E2PPi) formed by inorganic phosphate (Pi) and Mg2+ in the backward reaction, with and without ouabain or istaroxime, representatives of classical and new-generation cardiotonic steroids (CTSs). These two E2P states exhibit different biochemical properties. In particular, K+-sensitive acceleration of the dephosphorylation reaction is not observed with E2PPi, attributed to the presence of a Mg2+ ion in the transmembrane cation binding sites. The cryo-EM structures of NKA demonstrate that the two E2P structures are nearly identical but Mg2+ in the transmembrane binding cavity is identified only in E2PPi, corroborating the idea that it should be denoted as E2PPi·Mg2+. We can now explain why the absence of transmembrane Mg2+ in E2PATP confers the K+ sensitivity in dephosphorylation. In addition, we show that ATP bridges the actuator (A) and nucleotide binding (N) domains, stabilizing the E2PATP state; CTS binding causes hardly any changes in the structure of NKA, both in E2PATP and E2PPi·Mg2+, indicating that the binding mechanism is conformational selection; and istaroxime binds to NKA, extending its aminoalkyloxime group deep into the cation binding site. This orientation is upside down compared to that of classical CTSs with respect to the steroid ring. Notably, mobile parts of NKA are resolved substantially better in the electron microscopy (EM) maps than in previous X-ray structures, including sugars sticking out from the β-subunit and many phospholipid molecules. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32900.map.gz emd_32900.map.gz | 48.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32900-v30.xml emd-32900-v30.xml emd-32900.xml emd-32900.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32900_fsc.xml emd_32900_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_32900.png emd_32900.png | 143.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32900 http://ftp.pdbj.org/pub/emdb/structures/EMD-32900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32900 | HTTPS FTP |
-Validation report
| Summary document |  emd_32900_validation.pdf.gz emd_32900_validation.pdf.gz | 520.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32900_full_validation.pdf.gz emd_32900_full_validation.pdf.gz | 519.9 KB | Display | |
| Data in XML |  emd_32900_validation.xml.gz emd_32900_validation.xml.gz | 10.5 KB | Display | |
| Data in CIF |  emd_32900_validation.cif.gz emd_32900_validation.cif.gz | 13.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32900 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32900 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32900 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32900 | HTTPS FTP |
-Related structure data
| Related structure data |  7wz0MC  7wysC  7wytC  7wyuC  7wyvC  7wywC  7wyxC  7wyyC  7wyzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32900.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32900.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.076 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Sodium/potassium-transporting ATPase
| Entire | Name: Sodium/potassium-transporting ATPase |
|---|---|
| Components |
|
-Supramolecule #1: Sodium/potassium-transporting ATPase
| Supramolecule | Name: Sodium/potassium-transporting ATPase / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Squalus acanthias (spiny dogfish) Squalus acanthias (spiny dogfish) |
-Macromolecule #1: Sodium/potassium-transporting ATPase subunit alpha
| Macromolecule | Name: Sodium/potassium-transporting ATPase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  spiny dogfish (spiny dogfish) spiny dogfish (spiny dogfish) |
| Molecular weight | Theoretical: 113.389859 KDa |
| Sequence | String: MGKGTASDKY EPAATSENAT KSKKKGKKDK IDKKRDLDEL KKEVSMDDHK LSLDELHNKY GTDLTRGLTN ARAKEILARD GPNSLTPPP TTPEWIKFCR QLFGGFSILL WIGAILCFLA YGIQAATEDE PANDNLYLGV VLSTVVIVTG CFSYYQEAKS S RIMDSFKN ...String: MGKGTASDKY EPAATSENAT KSKKKGKKDK IDKKRDLDEL KKEVSMDDHK LSLDELHNKY GTDLTRGLTN ARAKEILARD GPNSLTPPP TTPEWIKFCR QLFGGFSILL WIGAILCFLA YGIQAATEDE PANDNLYLGV VLSTVVIVTG CFSYYQEAKS S RIMDSFKN MVPQQALVIR DGEKSTINAE FVVAGDLVEV KGGDRIPADL RIISAHGCKV DNSSLTGESE PQTRSPEFSS EN PLETRNI AFFSTNCVEG TARGVVVYTG DRTVMGRIAT LASGLEVGRT PIAIEIEHFI HIITGVAVFL GVSFFILSLI LGY SWLEAV IFLIGIIVAN VPEGLLATVT VCLTLTAKRM ARKNCLVKNL EAVETLGSTS TICS(PHD)KTGTL TQNRMTVAHM WFDNQIHEA DTTENQSGAA FDKTSATWSA LSRIAALCNR AVFQAGQDNV PILKRSVAGD ASESALLKCI ELCCGSVQGM R DRNPKIVE IPFNSTNKYQ LSIHENEKSS ESRYLLVMKG APERILDRCS TILLNGAEEP LKEDMKEAFQ NAYLELGGLG ER VLGFCHF ALPEDKYNEG YPFDADEPNF PTTDLCFVGL MAMIDPPRAA VPDAVGKCRS AGIKVIMVTG DHPITAKAIA KGV GIISEG NETIEDIAAR LNIPIGQVNP RDAKACVVHG SDLKDLSTEV LDDILHYHTE IVFARTSPQQ KLIIVEGCQR QGAI VAVTG DGVNDSPALK KADIGVAMGI SGSDVSKQAA DMILLDDNFA SIVTGVEEGR LIFDNLKKSI AYTLTSNIPE ITPFL VFII GNVPLPLGTV TILCIDLGTD MVPAISLAYE QAESDIMKRQ PRNPKTDKLV NERLISMAYG QIGMIQALGG FFSYFV ILA ENGFLPMDLI GKRVRWDDRW ISDVEDSFGQ QWTYEQRKIV EFTCHTSFFI SIVVVQWADL IICKTRRNSI FQQGMKN KI LIFGLFEETA LAAFLSYCPG TDVALRMYPL KPSWWFCAFP YSLIIFLYDE MRRFIIRRSP GGWVEQETYY |
-Macromolecule #2: Na+,K+-ATPase beta subunit
| Macromolecule | Name: Na+,K+-ATPase beta subunit / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  spiny dogfish (spiny dogfish) spiny dogfish (spiny dogfish) |
| Molecular weight | Theoretical: 35.176125 KDa |
| Sequence | String: MARGKSKETD GGWKKFLWDS EKKEFLGRTG SSWFKIFLFY LIFYGCLAGI FIGTIQVLLL TLSDFEPKYQ DRVAPPGLSH APYAIKTEI SFSISNPKSY ESFVKSMHKL MDLYNESSQA GNSPFEDCSD TPADYIKRGD LDDSQGQKKA CRFSRMWLKN C SGLDDTTY ...String: MARGKSKETD GGWKKFLWDS EKKEFLGRTG SSWFKIFLFY LIFYGCLAGI FIGTIQVLLL TLSDFEPKYQ DRVAPPGLSH APYAIKTEI SFSISNPKSY ESFVKSMHKL MDLYNESSQA GNSPFEDCSD TPADYIKRGD LDDSQGQKKA CRFSRMWLKN C SGLDDTTY GYAEGKPCVV AKLNRIIGFY PKPLKNTTDL PEELQANYNQ YVLPLRCAAK REEDREKIGS IEYFGLGGYA GF PLQYYPY YGKRLQKKYL QPLLAIQFTN LTQNMELRIE CKVYGENIDY SEKDRFRGRF EVKIEVKS |
-Macromolecule #3: FXYD domain-containing ion transport regulator
| Macromolecule | Name: FXYD domain-containing ion transport regulator / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  spiny dogfish (spiny dogfish) spiny dogfish (spiny dogfish) |
| Molecular weight | Theoretical: 10.195847 KDa |
| Sequence | String: MLGAATGLMV LVAVTQGVWA MDPEGPDNDE RFTYDYYRLR VVGLIVAAVL CVIGIIILLA GKCRCKFNQN KRTRSNSGTA TAQHLLQPG EATEC |
-Macromolecule #8: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 8 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #9: OUABAIN
| Macromolecule | Name: OUABAIN / type: ligand / ID: 9 / Number of copies: 2 / Formula: OBN |
|---|---|
| Molecular weight | Theoretical: 584.652 Da |
| Chemical component information |  ChemComp-OBN: |
-Macromolecule #10: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 10 / Number of copies: 8 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #11: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 11 / Number of copies: 24 / Formula: PCW |
|---|---|
| Molecular weight | Theoretical: 787.121 Da |
| Chemical component information |  ChemComp-PCW: |
-Macromolecule #12: water
| Macromolecule | Name: water / type: ligand / ID: 12 / Number of copies: 10 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99.9 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)