[English] 日本語
 Yorodumi
Yorodumi- EMDB-32801: Cryo-EM structure of a human pre-40S ribosomal subunit - State RR... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A1 (without CK1) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome biogenesis / 40S ribosome / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidyl-glutamine methylation / regulation of protein localization to nucleolus / rRNA (guanine-N7)-methylation / tRNA methyltransferase activator activity / rRNA (guanine) methyltransferase activity / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / tRNA modification in the nucleus and cytosol / Methylation / trophectodermal cell differentiation / protein methyltransferase activity ...peptidyl-glutamine methylation / regulation of protein localization to nucleolus / rRNA (guanine-N7)-methylation / tRNA methyltransferase activator activity / rRNA (guanine) methyltransferase activity / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / tRNA modification in the nucleus and cytosol / Methylation / trophectodermal cell differentiation / protein methyltransferase activity / positive regulation of rRNA processing / tRNA methylation / positive regulation of respiratory burst involved in inflammatory response / nucleolus organization / negative regulation of RNA splicing / rRNA methylation / neural crest cell differentiation / U3 snoRNA binding / rRNA modification in the nucleus and cytosol / erythrocyte homeostasis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / preribosome, small subunit precursor / negative regulation of ubiquitin protein ligase activity / Ribosomal scanning and start codon recognition / snoRNA binding / Translation initiation complex formation / mammalian oogenesis stage / activation-induced cell death of T cells / fibroblast growth factor binding / monocyte chemotaxis / Protein hydroxylation / SARS-CoV-1 modulates host translation machinery / mTORC1-mediated signalling / Peptide chain elongation / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / Selenocysteine synthesis / positive regulation of signal transduction by p53 class mediator / Formation of a pool of free 40S subunits / ubiquitin ligase inhibitor activity / Eukaryotic Translation Termination / negative regulation of respiratory burst involved in inflammatory response / Response of EIF2AK4 (GCN2) to amino acid deficiency / SRP-dependent cotranslational protein targeting to membrane / Viral mRNA Translation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / 90S preribosome / L13a-mediated translational silencing of Ceruloplasmin expression / TOR signaling / T cell proliferation involved in immune response / regulation of translational fidelity / Major pathway of rRNA processing in the nucleolus and cytosol / erythrocyte development / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / negative regulation of ubiquitin-dependent protein catabolic process / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / ribosomal small subunit export from nucleus / positive regulation of cell cycle / Nuclear events stimulated by ALK signaling in cancer / translation regulator activity / stress granule assembly / Mitotic Prometaphase / rough endoplasmic reticulum / EML4 and NUDC in mitotic spindle formation / gastrulation / MDM2/MDM4 family protein binding / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Prevention of phagosomal-lysosomal fusion / IRAK2 mediated activation of TAK1 complex / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / Membrane binding and targetting of GAG proteins / Endosomal Sorting Complex Required For Transport (ESCRT) / translation initiation factor binding / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / TICAM1,TRAF6-dependent induction of TAK1 complex / Negative regulation of FLT3 / Constitutive Signaling by NOTCH1 HD Domain Mutants / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Resolution of Sister Chromatid Cohesion / TICAM1-dependent activation of IRF3/IRF7 / NOTCH2 Activation and Transmission of Signal to the Nucleus / cytosolic ribosome / Regulation of FZD by ubiquitination / APC/C:Cdc20 mediated degradation of Cyclin B / Downregulation of ERBB4 signaling / p75NTR recruits signalling complexes / APC-Cdc20 mediated degradation of Nek2A / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Regulation of innate immune responses to cytosolic DNA Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
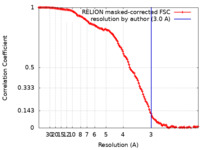
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Cheng J / Lau B / Thoms M / Ameismeier M / Berninghausen O / Hurt E / Beckmann R | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: The nucleoplasmic phase of pre-40S formation prior to nuclear export. Authors: Jingdong Cheng / Benjamin Lau / Matthias Thoms / Michael Ameismeier / Otto Berninghausen / Ed Hurt / Roland Beckmann /   Abstract: Biogenesis of the small ribosomal subunit in eukaryotes starts in the nucleolus with the formation of a 90S precursor and ends in the cytoplasm. Here, we elucidate the enigmatic structural ...Biogenesis of the small ribosomal subunit in eukaryotes starts in the nucleolus with the formation of a 90S precursor and ends in the cytoplasm. Here, we elucidate the enigmatic structural transitions of assembly intermediates from human and yeast cells during the nucleoplasmic maturation phase. After dissociation of all 90S factors, the 40S body adopts a close-to-mature conformation, whereas the 3' major domain, later forming the 40S head, remains entirely immature. A first coordination is facilitated by the assembly factors TSR1 and BUD23-TRMT112, followed by re-positioning of RRP12 that is already recruited early to the 90S for further head rearrangements. Eventually, the uS2 cluster, CK1 (Hrr25 in yeast) and the export factor SLX9 associate with the pre-40S to provide export competence. These exemplary findings reveal the evolutionary conserved mechanism of how yeast and humans assemble the 40S ribosomal subunit, but reveal also a few minor differences. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32801.map.gz emd_32801.map.gz | 106.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32801-v30.xml emd-32801-v30.xml emd-32801.xml emd-32801.xml | 49.7 KB 49.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32801_fsc.xml emd_32801_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_32801.png emd_32801.png | 155.1 KB | ||
| Filedesc metadata |  emd-32801.cif.gz emd-32801.cif.gz | 12.2 KB | ||
| Others |  emd_32801_additional_1.map.gz emd_32801_additional_1.map.gz emd_32801_additional_2.map.gz emd_32801_additional_2.map.gz | 105.6 MB 89.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32801 http://ftp.pdbj.org/pub/emdb/structures/EMD-32801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32801 | HTTPS FTP |
-Validation report
| Summary document |  emd_32801_validation.pdf.gz emd_32801_validation.pdf.gz | 454.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32801_full_validation.pdf.gz emd_32801_full_validation.pdf.gz | 454.2 KB | Display | |
| Data in XML |  emd_32801_validation.xml.gz emd_32801_validation.xml.gz | 13.2 KB | Display | |
| Data in CIF |  emd_32801_validation.cif.gz emd_32801_validation.cif.gz | 17.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32801 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32801 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32801 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32801 | HTTPS FTP |
-Related structure data
| Related structure data |  7wtuMC  7wtnC  7wtoC  7wtpC  7wtqC  7wtrC  7wtsC  7wttC  7wtvC  7wtwC  7wtxC  7wtzC  7wu0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32801.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32801.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
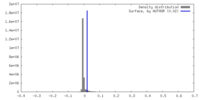
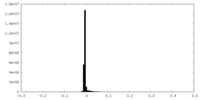
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #2
| File | emd_32801_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
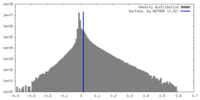
| Density Histograms |
-Additional map: #1
| File | emd_32801_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
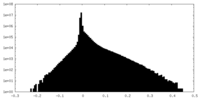
| Density Histograms |
- Sample components
Sample components
+Entire : Yeast pre-40S ribosomal subunit
+Supramolecule #1: Yeast pre-40S ribosomal subunit
+Macromolecule #1: 18S rRNA
+Macromolecule #2: 40S ribosomal protein S17
+Macromolecule #3: 40S ribosomal protein S27
+Macromolecule #4: 40S ribosomal protein S3a
+Macromolecule #5: 40S ribosomal protein S28
+Macromolecule #6: 40S ribosomal protein S4, X isoform
+Macromolecule #7: 40S ribosomal protein S30
+Macromolecule #8: 40S ribosomal protein S5
+Macromolecule #9: 40S ribosomal protein S7
+Macromolecule #10: 40S ribosomal protein S6
+Macromolecule #11: 40S ribosomal protein S25
+Macromolecule #12: 40S ribosomal protein S24
+Macromolecule #13: RNA-binding protein PNO1
+Macromolecule #14: 40S ribosomal protein S23
+Macromolecule #15: Bystin
+Macromolecule #16: 40S ribosomal protein S15a
+Macromolecule #17: Pre-rRNA-processing protein TSR1 homolog
+Macromolecule #18: Protein LTV1 homolog
+Macromolecule #19: 40S ribosomal protein S19
+Macromolecule #20: 40S ribosomal protein S18
+Macromolecule #21: 40S ribosomal protein S16
+Macromolecule #22: 40S ribosomal protein S15
+Macromolecule #23: 40S ribosomal protein S14
+Macromolecule #24: 40S ribosomal protein S13
+Macromolecule #25: 40S ribosomal protein S11
+Macromolecule #26: 40S ribosomal protein S9
+Macromolecule #27: 40S ribosomal protein S8
+Macromolecule #28: Multifunctional methyltransferase subunit TRM112-like protein
+Macromolecule #29: Probable 18S rRNA (guanine-N(7))-methyltransferase
+Macromolecule #30: RRP12-like protein
+Macromolecule #31: 40S ribosomal protein S12
+Macromolecule #32: Ubiquitin-40S ribosomal protein S27a
+Macromolecule #33: Ribosome biogenesis protein SLX9 homolog
+Macromolecule #34: S-ADENOSYL-L-HOMOCYSTEINE
+Macromolecule #35: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)