[English] 日本語
 Yorodumi
Yorodumi- EMDB-32139: Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32139 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80 (local) | |||||||||
 Map data Map data | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, local map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G Protein-Coupled Receptor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmast cell secretagogue receptor activity / mast cell activation / sleep / neuropeptide binding / mast cell degranulation / positive regulation of cytokinesis / sensory perception of pain / G protein-coupled receptor activity / G protein-coupled receptor signaling pathway / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Li Y / Yang F | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structure, function and pharmacology of human itch receptor complexes. Authors: Fan Yang / Lulu Guo / Yu Li / Guopeng Wang / Jia Wang / Chao Zhang / Guo-Xing Fang / Xu Chen / Lei Liu / Xu Yan / Qun Liu / Changxiu Qu / Yunfei Xu / Peng Xiao / Zhongliang Zhu / Zijian Li / ...Authors: Fan Yang / Lulu Guo / Yu Li / Guopeng Wang / Jia Wang / Chao Zhang / Guo-Xing Fang / Xu Chen / Lei Liu / Xu Yan / Qun Liu / Changxiu Qu / Yunfei Xu / Peng Xiao / Zhongliang Zhu / Zijian Li / Jiuyao Zhou / Xiao Yu / Ning Gao / Jin-Peng Sun /  Abstract: In the clades of animals that diverged from the bony fish, a group of Mas-related G-protein-coupled receptors (MRGPRs) evolved that have an active role in itch and allergic signals. As an MRGPR, ...In the clades of animals that diverged from the bony fish, a group of Mas-related G-protein-coupled receptors (MRGPRs) evolved that have an active role in itch and allergic signals. As an MRGPR, MRGPRX2 is known to sense basic secretagogues (agents that promote secretion) and is involved in itch signals and eliciting pseudoallergic reactions. MRGPRX2 has been targeted by drug development efforts to prevent the side effects induced by certain drugs or to treat allergic diseases. Here we report a set of cryo-electron microscopy structures of the MRGPRX2-G trimer in complex with polycationic compound 48/80 or with inflammatory peptides. The structures of the MRGPRX2-G complex exhibited shallow, solvent-exposed ligand-binding pockets. We identified key common structural features of MRGPRX2 and describe a consensus motif for peptidic allergens. Beneath the ligand-binding pocket, the unusual kink formation at transmembrane domain 6 (TM6) and the replacement of the general toggle switch from Trp to Gly (superscript annotations as per Ballesteros-Weinstein nomenclature) suggest a distinct activation process. We characterized the interfaces of MRGPRX2 and the G trimer, and mapped the residues associated with key single-nucleotide polymorphisms on both the ligand and G-protein interfaces of MRGPRX2. Collectively, our results provide a structural basis for the sensing of cationic allergens by MRGPRX2, potentially facilitating the rational design of therapies to prevent unwanted pseudoallergic reactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32139.map.gz emd_32139.map.gz | 541.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32139-v30.xml emd-32139-v30.xml emd-32139.xml emd-32139.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32139.png emd_32139.png | 40.1 KB | ||
| Filedesc metadata |  emd-32139.cif.gz emd-32139.cif.gz | 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32139 http://ftp.pdbj.org/pub/emdb/structures/EMD-32139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32139 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32139 | HTTPS FTP |
-Validation report
| Summary document |  emd_32139_validation.pdf.gz emd_32139_validation.pdf.gz | 343.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32139_full_validation.pdf.gz emd_32139_full_validation.pdf.gz | 343.3 KB | Display | |
| Data in XML |  emd_32139_validation.xml.gz emd_32139_validation.xml.gz | 5.3 KB | Display | |
| Data in CIF |  emd_32139_validation.cif.gz emd_32139_validation.cif.gz | 6.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32139 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32139 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32139 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32139 | HTTPS FTP |
-Related structure data
| Related structure data |  7vv6MC  7vdhC  7vdlC  7vdmC  7vuyC  7vuzC  7vv0C  7vv3C  7vv4C  7vv5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32139.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32139.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, local map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.052 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with...
| Entire | Name: Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80 (local) |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with...
| Supramolecule | Name: Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80 (local) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Mas-related G-protein coupled receptor member X2
| Macromolecule | Name: Mas-related G-protein coupled receptor member X2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.123984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDPTTPAWGT ESTTVNGNDQ ALLLLCGKET LIPVFLILFI ALVGLVGNGF VLWLLGFRMR RNAFSVYVLS LAGADFLFLC FQIINCLVY LSNFFCSISI NFPSFFTTVM TCAYLAGLSM LSTVSTERCL SVLWPIWYRC RRPRHLSAVV CVLLWALSLL L SILEGKFC ...String: MDPTTPAWGT ESTTVNGNDQ ALLLLCGKET LIPVFLILFI ALVGLVGNGF VLWLLGFRMR RNAFSVYVLS LAGADFLFLC FQIINCLVY LSNFFCSISI NFPSFFTTVM TCAYLAGLSM LSTVSTERCL SVLWPIWYRC RRPRHLSAVV CVLLWALSLL L SILEGKFC GFLFSDGDSG WCQTFDFITA AWLIFLFMVL CGSSLALLVR ILCGSRGLPL TRLYLTILLT VLVFLLCGLP FG IQWFLIL WIWKDSDVLF CHIHPVSVVL SSLNSSANPI IYFFVGSFRK QWRLQQPILK LALQRALQDI AEVDHSEGCF RQG TPEMSR SSLV UniProtKB: Mas-related G-protein coupled receptor member X2 |
-Macromolecule #2: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 2 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #3: 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]...
| Macromolecule | Name: 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine type: ligand / ID: 3 / Number of copies: 1 / Formula: 6IB |
|---|---|
| Molecular weight | Theoretical: 519.718 Da |
| Chemical component information | 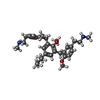 ChemComp-6IB: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Number images used: 587441 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















