[English] 日本語
 Yorodumi
Yorodumi- EMDB-30932: LHCII-1 in the state transition supercomplex PSI-LHCI-LHCII from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30932 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | LHCII-1 in the state transition supercomplex PSI-LHCI-LHCII from the LhcbM1 lacking mutant of Chlamydomonas reinhardtii | |||||||||||||||||||||||||||
 Map data Map data | Local refined map of LHCII-1 in the PSI-LHCI-LHCII supercomplex from LhcbM1 lacking mutant of Chlamydomonas reinhardtii | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | Supercomplex / LHCII / state transition / green alga / Chlamydomonas reinhardtii / PHOTOSYNTHESIS | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthesis, light harvesting in photosystem I / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane / response to light stimulus Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.75 Å | |||||||||||||||||||||||||||
 Authors Authors | Pan XW / Li AJ | |||||||||||||||||||||||||||
| Funding support |  China, China,  Japan, 8 items Japan, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2021 Journal: Nat Plants / Year: 2021Title: Structural basis of LhcbM5-mediated state transitions in green algae. Authors: Xiaowei Pan / Ryutaro Tokutsu / Anjie Li / Kenji Takizawa / Chihong Song / Kazuyoshi Murata / Tomohito Yamasaki / Zhenfeng Liu / Jun Minagawa / Mei Li /   Abstract: In green algae and plants, state transitions serve as a short-term light-acclimation process in the regulation of the light-harvesting capacity of photosystems I and II (PSI and PSII, respectively). ...In green algae and plants, state transitions serve as a short-term light-acclimation process in the regulation of the light-harvesting capacity of photosystems I and II (PSI and PSII, respectively). During the process, a portion of light-harvesting complex II (LHCII) is phosphorylated, dissociated from PSII and binds with PSI to form the supercomplex PSI-LHCI-LHCII. Here, we report high-resolution structures of PSI-LHCI-LHCII from Chlamydomonas reinhardtii, revealing the mechanism of assembly between the PSI-LHCI complex and two phosphorylated LHCII trimers containing all four types of LhcbM protein. Two specific LhcbM isoforms, namely LhcbM1 and LhcbM5, directly interact with the PSI core through their phosphorylated amino terminal regions. Furthermore, biochemical and functional studies on mutant strains lacking either LhcbM1 or LhcbM5 indicate that only LhcbM5 is indispensable in supercomplex formation. The results unravel the specific interactions and potential excitation energy transfer routes between green algal PSI and two phosphorylated LHCIIs. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30932.map.gz emd_30932.map.gz | 5.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30932-v30.xml emd-30932-v30.xml emd-30932.xml emd-30932.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30932.png emd_30932.png | 47.4 KB | ||
| Filedesc metadata |  emd-30932.cif.gz emd-30932.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30932 http://ftp.pdbj.org/pub/emdb/structures/EMD-30932 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30932 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30932 | HTTPS FTP |
-Validation report
| Summary document |  emd_30932_validation.pdf.gz emd_30932_validation.pdf.gz | 323.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30932_full_validation.pdf.gz emd_30932_full_validation.pdf.gz | 322.9 KB | Display | |
| Data in XML |  emd_30932_validation.xml.gz emd_30932_validation.xml.gz | 5.7 KB | Display | |
| Data in CIF |  emd_30932_validation.cif.gz emd_30932_validation.cif.gz | 6.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30932 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30932 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30932 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30932 | HTTPS FTP |
-Related structure data
| Related structure data |  7e0hMC  7dz7C  7dz8C  7e0iC  7e0jC  7e0kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30932.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30932.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refined map of LHCII-1 in the PSI-LHCI-LHCII supercomplex from LhcbM1 lacking mutant of Chlamydomonas reinhardtii | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
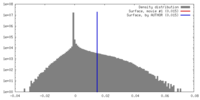
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : State transition complex PSI-LHCI-LHCII from green algae
| Entire | Name: State transition complex PSI-LHCI-LHCII from green algae |
|---|---|
| Components |
|
-Supramolecule #1: State transition complex PSI-LHCI-LHCII from green algae
| Supramolecule | Name: State transition complex PSI-LHCI-LHCII from green algae type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Chlorophyll a-b binding protein, chloroplastic
| Macromolecule | Name: Chlorophyll a-b binding protein, chloroplastic / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.67535 KDa |
| Sequence | String: MAAIMKSAVR SSVRPTVSGR SARVVPRAAI EWYGPDRPKF LGPFSEGDTP AYLTGEFPGD YGWDTAGLSA DPETFKRYRE LELIHARWA MLGALGCITP ELLAKNGIPF GEAVWFKAGA QIFAEGGLNY LGNENLIHAQ SIIATLAFQV VVMGLAEAYR A NGGPLGEG ...String: MAAIMKSAVR SSVRPTVSGR SARVVPRAAI EWYGPDRPKF LGPFSEGDTP AYLTGEFPGD YGWDTAGLSA DPETFKRYRE LELIHARWA MLGALGCITP ELLAKNGIPF GEAVWFKAGA QIFAEGGLNY LGNENLIHAQ SIIATLAFQV VVMGLAEAYR A NGGPLGEG LDPLHPGGAF DPLGLADDPD TFAELKVKEI KNGRLAMFSM FGFFVQAIVT GKGPIQNLDD HLANPTAVNA FA YATKFTP SA UniProtKB: Chlorophyll a-b binding protein, chloroplastic |
-Macromolecule #2: Chlorophyll a-b binding protein, chloroplastic
| Macromolecule | Name: Chlorophyll a-b binding protein, chloroplastic / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.405008 KDa |
| Sequence | String: MAFALSFSRK ALQVSAKATG KKGTGKTAAK QAPASSGIEF YGPNRAKWLG PYSENATPAY LTGEFPGDYG WDTAGLSADP ETFKRYREL ELIHARWAML GALGCITPEL LAKSGTQFGE AVWFKAGAQI FSEGGLDYLG NPSLVHAQNI VATLAVQVIL M GLVEGYRV ...String: MAFALSFSRK ALQVSAKATG KKGTGKTAAK QAPASSGIEF YGPNRAKWLG PYSENATPAY LTGEFPGDYG WDTAGLSADP ETFKRYREL ELIHARWAML GALGCITPEL LAKSGTQFGE AVWFKAGAQI FSEGGLDYLG NPSLVHAQNI VATLAVQVIL M GLVEGYRV NGGPAGEGLD PLYPGESFDP LGLADDPDTF AELKVKEIKN GRLAMFSMFG FFVQAIVTGK GPIQNLDDHL SN PTVNNAF AFATKFTPSA UniProtKB: Chlorophyll a-b binding protein, chloroplastic |
-Macromolecule #3: CHLOROPHYLL B
| Macromolecule | Name: CHLOROPHYLL B / type: ligand / ID: 3 / Number of copies: 18 / Formula: CHL |
|---|---|
| Molecular weight | Theoretical: 907.472 Da |
| Chemical component information |  ChemComp-CHL: |
-Macromolecule #4: CHLOROPHYLL A
| Macromolecule | Name: CHLOROPHYLL A / type: ligand / ID: 4 / Number of copies: 24 / Formula: CLA |
|---|---|
| Molecular weight | Theoretical: 893.489 Da |
| Chemical component information |  ChemComp-CLA: |
-Macromolecule #5: (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL
| Macromolecule | Name: (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL type: ligand / ID: 5 / Number of copies: 6 / Formula: LUT |
|---|---|
| Molecular weight | Theoretical: 568.871 Da |
| Chemical component information |  ChemComp-LUT: |
-Macromolecule #6: (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BE...
| Macromolecule | Name: (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL type: ligand / ID: 6 / Number of copies: 3 / Formula: XAT |
|---|---|
| Molecular weight | Theoretical: 600.87 Da |
| Chemical component information |  ChemComp-XAT: |
-Macromolecule #7: (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY...
| Macromolecule | Name: (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL type: ligand / ID: 7 / Number of copies: 3 / Formula: NEX |
|---|---|
| Molecular weight | Theoretical: 600.87 Da |
| Chemical component information |  ChemComp-NEX: |
-Macromolecule #8: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
| Macromolecule | Name: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE / type: ligand / ID: 8 / Number of copies: 3 / Formula: LHG |
|---|---|
| Molecular weight | Theoretical: 722.97 Da |
| Chemical component information |  ChemComp-LHG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.5625 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.75 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 123997 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)