[English] 日本語
 Yorodumi
Yorodumi- PDB-2bq2: Solution Structure of the DNA Duplex ACGCGU-NA with a 2' Amido-Li... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2bq2 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Solution Structure of the DNA Duplex ACGCGU-NA with a 2' Amido-Linked Nalidixic Acid Residue at the 3' Terminal Nucleotide | |||||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / NUCLEIC ACID / DEOXYRIBONUCLEIC ACID / DISRUPTED TERMINAL BASEPAIRS / QUINOLONE / SYNTHETIC HYBRID / DNA COMPLEX | Function / homology | NALIDIXIC ACID / DNA |  Function and homology information Function and homology informationMethod | SOLUTION NMR / simulated annealing |  Authors AuthorsSiegmund, K. / Maheshwary, S. / Narayanan, S. / Connors, W. / Richert, M. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2005 Journal: Nucleic Acids Res. / Year: 2005Title: Molecular details of quinolone-DNA interactions: solution structure of an unusually stable DNA duplex with covalently linked nalidixic acid residues and non-covalent complexes derived from it. Authors: Siegmund, K. / Maheshwary, S. / Narayanan, S. / Connors, W. / Riedrich, M. / Printz, M. / Richert, C. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2bq2.cif.gz 2bq2.cif.gz | 26.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2bq2.ent.gz pdb2bq2.ent.gz | 19 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2bq2.json.gz 2bq2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2bq2_validation.pdf.gz 2bq2_validation.pdf.gz | 328.3 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2bq2_full_validation.pdf.gz 2bq2_full_validation.pdf.gz | 333 KB | Display | |
| Data in XML |  2bq2_validation.xml.gz 2bq2_validation.xml.gz | 1.9 KB | Display | |
| Data in CIF |  2bq2_validation.cif.gz 2bq2_validation.cif.gz | 2.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bq/2bq2 https://data.pdbj.org/pub/pdb/validation_reports/bq/2bq2 ftp://data.pdbj.org/pub/pdb/validation_reports/bq/2bq2 ftp://data.pdbj.org/pub/pdb/validation_reports/bq/2bq2 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 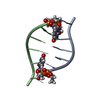
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 1810.206 Da / Num. of mol.: 2 / Source method: obtained synthetically Details: NALIDIXIC ACID APPENDED TO THE AMINO FUNCTION OF 2'-AMINOURIDINE VIA AN AMIDE BOND #2: Chemical | |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: NONE |
- Sample preparation
Sample preparation
| Details | Contents: D2O AND 90% WATER/ 10%D2O |
|---|---|
| Sample conditions | Ionic strength: 160 mM / pH: 7 / Pressure: 1.0 atm / Temperature: 283.0 K |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE / Manufacturer: Bruker / Model: AVANCE / Field strength: 800 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 / Details: SEE PRIMARY CITATION FOR DETAILS | |||||||||
| NMR ensemble | Conformer selection criteria: LOWEST CALCULATED ENERGIES / Conformers calculated total number: 80 / Conformers submitted total number: 2 |
 Movie
Movie Controller
Controller






 PDBj
PDBj



 HSQC
HSQC