[English] 日本語
 Yorodumi
Yorodumi- PDB-1c0o: SOLUTION STRUCTURE OF THE P5 HAIRPIN FROM A GROUP I INTRON COMPLE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1c0o | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF THE P5 HAIRPIN FROM A GROUP I INTRON COMPLEXED WITH COBALT (III) HEXAMMINE, NMR, 19 CONVERGED STRUCTURES | ||||||||||||||||||||
 Components Components | RNA (5'-R(* Keywords KeywordsRNA / RIBONUCLEIC ACID / COBALT (III) HEXAMMINE / METAL BINDING / RNA STRUCTURE | Function / homology | COBALT HEXAMMINE(III) / RNA / RNA (> 10) |  Function and homology information Function and homology informationBiological species |  Method | SOLUTION NMR / RESTRAINED MOLECULAR DYNAMICS |  Authors AuthorsColmenarejo, G. / Tinoco Jr., I. |  Citation Citation Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: Structure and thermodynamics of metal binding in the P5 helix of a group I intron ribozyme. Authors: Colmenarejo, G. / Tinoco Jr., I. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1c0o.cif.gz 1c0o.cif.gz | 176.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1c0o.ent.gz pdb1c0o.ent.gz | 146.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1c0o.json.gz 1c0o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1c0o_validation.pdf.gz 1c0o_validation.pdf.gz | 335.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1c0o_full_validation.pdf.gz 1c0o_full_validation.pdf.gz | 419.1 KB | Display | |
| Data in XML |  1c0o_validation.xml.gz 1c0o_validation.xml.gz | 5 KB | Display | |
| Data in CIF |  1c0o_validation.cif.gz 1c0o_validation.cif.gz | 8.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c0/1c0o https://data.pdbj.org/pub/pdb/validation_reports/c0/1c0o ftp://data.pdbj.org/pub/pdb/validation_reports/c0/1c0o ftp://data.pdbj.org/pub/pdb/validation_reports/c0/1c0o | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 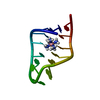
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 4471.667 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: CLOSED BY A UUCG TETRALOOP; COBALT (III) HEXAMMINE BINDS AT THE MAJOR GROOVE OF TWO TANDEM G-U WOBBLE PAIRS IN 5'-GU-3' ORIENTATION Source: (gene. exp.)  Description: SYNTHESIZED ENZYMATICALLY IN-VITRO USING T7 RNA POLYMERASE |
|---|---|
| #2: Chemical | ChemComp-NCO / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING A NON-ISOTOPICALLY LABELLED RNA SAMPLE. ALL 13C EXPERIMENTS WERE DONE USING NATURAL ABUNDANCE. COBALT (III) HEXAMMINE WAS LOCATED USING DOCKING WITH 7 ...Text: THE STRUCTURE WAS DETERMINED USING A NON-ISOTOPICALLY LABELLED RNA SAMPLE. ALL 13C EXPERIMENTS WERE DONE USING NATURAL ABUNDANCE. COBALT (III) HEXAMMINE WAS LOCATED USING DOCKING WITH 7 INTERMOLECULAR NOES OBSERVED IN 2D WATER NOESY EXPERIMENTS. |
- Sample preparation
Sample preparation
| Details | Contents: PHOSPHATE BUFFER, EDTA |
|---|---|
| Sample conditions | Ionic strength: 100 mM NACL / pH: 6.4 / Pressure: 1 atm / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: RESTRAINED MOLECULAR DYNAMICS / Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: NOE ENERGY (LEAST RESTRAINT VIOLATIONS), DIHEDRAL ENERGY (LEAST RESTRAINT VIOLATIONS), TOTAL ENERGY Conformers calculated total number: 40 / Conformers submitted total number: 19 |
 Movie
Movie Controller
Controller





 PDBj
PDBj































 X-PLOR
X-PLOR