[English] 日本語
 Yorodumi
Yorodumi- EMDB-2993: Structural rearrangements in the phage head-to-tail interface dur... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2993 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural rearrangements in the phage head-to-tail interface during assembly and infection | |||||||||
 Map data Map data | Structure of bacteriophage SPP1 head-to-tail interface filled with DNA and tape measure protein. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | viral infection / tailed bacteriophage / Siphoviridae / SPP1 / viral assembly / head-to-tail interface / DNA gatekeeper / allosteric mechanism / concerted reorganisation / diaphragm gating | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral head-tail joining / virus tail fiber assembly / viral DNA genome packaging, headful / virus tail, tube / viral procapsid / viral portal complex / symbiont genome ejection through host cell envelope, long flexible tail mechanism / virus tail / virion component / viral translational frameshifting Similarity search - Function | |||||||||
| Biological species |  Bacillus phage SPP1 (virus) Bacillus phage SPP1 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.6 Å | |||||||||
 Authors Authors | Chaban Y / Lurz R / Brasiles S / Cornilleau C / Karreman M / Zinn-Justin S / Tavares P / Orlova EV | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2015 Journal: Proc Natl Acad Sci U S A / Year: 2015Title: Structural rearrangements in the phage head-to-tail interface during assembly and infection. Authors: Yuriy Chaban / Rudi Lurz / Sandrine Brasilès / Charlène Cornilleau / Matthia Karreman / Sophie Zinn-Justin / Paulo Tavares / Elena V Orlova /    Abstract: Many icosahedral viruses use a specialized portal vertex to control genome encapsidation and release from the viral capsid. In tailed bacteriophages, the portal system is connected to a tail ...Many icosahedral viruses use a specialized portal vertex to control genome encapsidation and release from the viral capsid. In tailed bacteriophages, the portal system is connected to a tail structure that provides the pipeline for genome delivery to the host cell. We report the first, to our knowledge, subnanometer structures of the complete portal-phage tail interface that mimic the states before and after DNA release during phage infection. They uncover structural rearrangements associated with intimate protein-DNA interactions. The portal protein gp6 of bacteriophage SPP1 undergoes a concerted reorganization of the structural elements of its central channel during interaction with DNA. A network of protein-protein interactions primes consecutive binding of proteins gp15 and gp16 to extend and close the channel. This critical step that prevents genome leakage from the capsid is achieved by a previously unidentified allosteric mechanism: gp16 binding to two different regions of gp15 drives correct positioning and folding of an inner gp16 loop to interact with equivalent loops of the other gp16 subunits. Together, these loops build a plug that closes the channel. Gp16 then fastens the tail to yield the infectious virion. The gatekeeper system opens for viral genome exit at the beginning of infection but recloses afterward, suggesting a molecular diaphragm-like mechanism to control DNA efflux. The mechanisms described here, controlling the essential steps of phage genome movements during virus assembly and infection, are likely to be conserved among long-tailed phages, the largest group of viruses in the Biosphere. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2993.map.gz emd_2993.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2993-v30.xml emd-2993-v30.xml emd-2993.xml emd-2993.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| Images |  Spp1_EMD2993_500.jpg Spp1_EMD2993_500.jpg | 47 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2993 http://ftp.pdbj.org/pub/emdb/structures/EMD-2993 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2993 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2993 | HTTPS FTP |
-Related structure data
| Related structure data |  5a20MC  2994C  5a21C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2993.map.gz / Format: CCP4 / Size: 51.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2993.map.gz / Format: CCP4 / Size: 51.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of bacteriophage SPP1 head-to-tail interface filled with DNA and tape measure protein. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
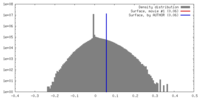
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacteriophage SPP1 head-to-tail interface
| Entire | Name: Bacteriophage SPP1 head-to-tail interface |
|---|---|
| Components |
|
-Supramolecule #1000: Bacteriophage SPP1 head-to-tail interface
| Supramolecule | Name: Bacteriophage SPP1 head-to-tail interface / type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: cyclical dodecamers and hexamers / Number unique components: 5 |
|---|---|
| Molecular weight | Theoretical: 1.2 MDa |
-Macromolecule #1: Bacteriophage SPP1 portal protein gp6
| Macromolecule | Name: Bacteriophage SPP1 portal protein gp6 / type: protein_or_peptide / ID: 1 / Name.synonym: gp6 / Number of copies: 12 / Oligomeric state: cyclical dodecamer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 |
| Molecular weight | Experimental: 57.2 KDa / Theoretical: 57.3225 KDa |
| Sequence | UniProtKB: Portal protein / InterPro: Portal protein, SPP1-type |
-Macromolecule #2: head completion protein gp15
| Macromolecule | Name: head completion protein gp15 / type: protein_or_peptide / ID: 2 / Name.synonym: gp15 / Number of copies: 12 / Oligomeric state: cyclical dodecamer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 |
| Molecular weight | Experimental: 11.6 KDa / Theoretical: 11.6144 KDa |
| Sequence | UniProtKB: Head completion protein gp15 / InterPro: Phage gp6-like head-tail connector protein |
-Macromolecule #3: head completion protein gp16
| Macromolecule | Name: head completion protein gp16 / type: protein_or_peptide / ID: 3 / Name.synonym: gp16 / Number of copies: 12 / Oligomeric state: cyclical dodecamer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 |
| Molecular weight | Experimental: 12.5 KDa / Theoretical: 12.542 KDa |
| Sequence | UniProtKB: Head completion protein gp16 / InterPro: Bacteriophage SPP1, head-tail adaptor |
-Macromolecule #4: tail-to-head joining protein gp17
| Macromolecule | Name: tail-to-head joining protein gp17 / type: protein_or_peptide / ID: 4 / Name.synonym: gp17 / Number of copies: 6 / Oligomeric state: cyclical hexamer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 |
| Molecular weight | Experimental: 15 KDa / Theoretical: 15.01 KDa |
| Sequence | UniProtKB: Tail completion protein gp17 / InterPro: Tail completion protein |
-Macromolecule #5: major tail protein gp17.1
| Macromolecule | Name: major tail protein gp17.1 / type: protein_or_peptide / ID: 5 / Name.synonym: gp17.1 / Number of copies: 6 / Oligomeric state: cyclical hexamer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 Bacillus phage SPP1 (virus) / Strain: Bacillus phage SPP1 / synonym: bacteriophage SPP1 |
| Molecular weight | Experimental: 19.2 KDa / Theoretical: 19.1158 KDa |
| Sequence | UniProtKB: Tail tube protein gp17.1* / InterPro: Phage major tail protein TP901-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM NaCl, 50 mM Tris-Cl, 5 mM MgCl2 |
|---|---|
| Grid | Details: holey carbon coated Quantifoil grids (r2/2) (Quantifoil, Germany) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK II / Method: blot for 2 seconds |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Date | Oct 9, 2008 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PRIMESCAN / Digitization - Sampling interval: 4 µm / Number real images: 200 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.6 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 39000 |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 7.6 Å / Resolution method: OTHER / Software - Name: Imagic, Spider, EMAN / Number images used: 14000 |
| Final two d classification | Number classes: 3500 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: Chimera, VEDA (UROX), Modeller Flex-EM |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-5a20: |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name: Chimera, VEDA (UROX), Modeller Flex-EM |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-5a20: |
-Atomic model buiding 3
| Initial model | PDB ID: |
|---|---|
| Software | Name: Chimera, VEDA (UROX), Modeller Flex-EM |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-5a20: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)