[English] 日本語
 Yorodumi
Yorodumi- EMDB-2984: 2.2 A resolution cryo-EM structure of beta-galactosidase in compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2984 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | |||||||||
 Map data Map data | B-factor corrected reconstruction of PETG-bound beta-galactosidase. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | near-atomic resolution cryo-electron microscopy / single-particle cryo-EM / protein complexes / PETG | |||||||||
| Function / homology |  Function and homology information Function and homology informationalkali metal ion binding / lactose catabolic process / beta-galactosidase complex / beta-galactosidase / beta-galactosidase activity / carbohydrate binding / magnesium ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
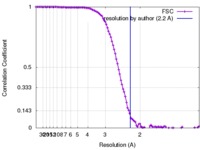
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Bartesaghi A / Merk A / Banerjee S / Matthies D / Wu X / Milne J / Subramaniam S | |||||||||
 Citation Citation |  Journal: Science / Year: 2015 Journal: Science / Year: 2015Title: 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Authors: Alberto Bartesaghi / Alan Merk / Soojay Banerjee / Doreen Matthies / Xiongwu Wu / Jacqueline L S Milne / Sriram Subramaniam /  Abstract: Cryo-electron microscopy (cryo-EM) is rapidly emerging as a powerful tool for protein structure determination at high resolution. Here we report the structure of a complex between Escherichia coli β- ...Cryo-electron microscopy (cryo-EM) is rapidly emerging as a powerful tool for protein structure determination at high resolution. Here we report the structure of a complex between Escherichia coli β-galactosidase and the cell-permeant inhibitor phenylethyl β-D-thiogalactopyranoside (PETG), determined by cryo-EM at an average resolution of ~2.2 angstroms (Å). Besides the PETG ligand, we identified densities in the map for ~800 water molecules and for magnesium and sodium ions. Although it is likely that continued advances in detector technology may further enhance resolution, our findings demonstrate that preparation of specimens of adequate quality and intrinsic protein flexibility, rather than imaging or image-processing technologies, now represent the major bottlenecks to routinely achieving resolutions close to 2 Å using single-particle cryo-EM. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2984.map.gz emd_2984.map.gz | 86.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2984-v30.xml emd-2984-v30.xml emd-2984.xml emd-2984.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2984_fsc.xml emd_2984_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd-2984-bgal2015_EMDB02.jpg emd-2984-bgal2015_EMDB02.jpg | 224.5 KB | ||
| Others |  emd_2984_additional_1.map.gz emd_2984_additional_1.map.gz emd_2984_half_map_1.map.gz emd_2984_half_map_1.map.gz emd_2984_half_map_2.map.gz emd_2984_half_map_2.map.gz | 87.3 MB 87.9 MB 87.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2984 http://ftp.pdbj.org/pub/emdb/structures/EMD-2984 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2984 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2984 | HTTPS FTP |
-Related structure data
| Related structure data |  5a1aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10061 (Title: 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor EMPIAR-10061 (Title: 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitorData size: 12.4 TB Data #1: Averages of aligned movie frames [micrographs - single frame] Data #2: Raw movie frames [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2984.map.gz / Format: CCP4 / Size: 92.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2984.map.gz / Format: CCP4 / Size: 92.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor corrected reconstruction of PETG-bound beta-galactosidase. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.637 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 2984 additional 1.map
| File | emd_2984_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 2984 half map 1.map
| File | emd_2984_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 2984 half map 2.map
| File | emd_2984_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia coli beta-galactosidase bound to phenylethyl beta-D-t...
| Entire | Name: Escherichia coli beta-galactosidase bound to phenylethyl beta-D-thiogalactopyranoside (PETG) |
|---|---|
| Components |
|
-Supramolecule #1000: Escherichia coli beta-galactosidase bound to phenylethyl beta-D-t...
| Supramolecule | Name: Escherichia coli beta-galactosidase bound to phenylethyl beta-D-thiogalactopyranoside (PETG) type: sample / ID: 1000 / Details: The sample was monodisperse. / Oligomeric state: tetramer / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 465 KDa Method: ProtParam tool: Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A., Protein Identification and Analysis Tools on the ExPASy Server, (in) John M. Walker ...Method: ProtParam tool: Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A., Protein Identification and Analysis Tools on the ExPASy Server, (in) John M. Walker (ed): The Proteomics Protocols Handbook, Humana Press (2005), pp. 571-607 |
-Macromolecule #1: beta-galactosidase
| Macromolecule | Name: beta-galactosidase / type: protein_or_peptide / ID: 1 / Name.synonym: beta-gal, b-gal / Number of copies: 1 / Oligomeric state: tetramer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 465 KDa |
| Sequence | UniProtKB: Beta-galactosidase / GO: beta-galactosidase activity |
-Macromolecule #2: phenylethyl beta-D-thiogalactopyranoside
| Macromolecule | Name: phenylethyl beta-D-thiogalactopyranoside / type: ligand / ID: 2 / Name.synonym: PETG / Details: Molecular weight of PETG is 300.37 Da. / Number of copies: 4 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.3 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 25 mM Tris, pH 8.0, 50 mM NaCl, 2 mM MgCl2, 1.0 mM TCEP |
| Grid | Details: 200 mesh Quantifoil R2/2 grids, plasma cleaned |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 90.15 K / Instrument: LEICA EM GP / Method: Blot for 2 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 79.6 K / Max: 79.8 K / Average: 79.7 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 215,000 times magnification. |
| Specialist optics | Energy filter - Name: Gatan, Inc. / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Details | Parallel beam illumination |
| Date | Dec 15, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 1487 / Average electron dose: 45 e/Å2 Details: Every image is the average of 38 frames recorded by the direct electron detector. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 215000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder: Liquid nitrogen cooled / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)