+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HMGCR-UBIAD1-BRIL-Fab-Nb Complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Chen H / Qi X / Li X | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain. Authors: Hongwen Chen / Xiaofeng Qi / Rebecca A Faulkner / Marc M Schumacher / Linda M Donnelly / Russell A DeBose-Boyd / Xiaochun Li /  Abstract: 3-Hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) is the rate-limiting enzyme in cholesterol synthesis and target of cholesterol-lowering statin drugs. Accumulation of sterols in endoplasmic ...3-Hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) is the rate-limiting enzyme in cholesterol synthesis and target of cholesterol-lowering statin drugs. Accumulation of sterols in endoplasmic reticulum (ER) membranes accelerates degradation of HMGCR, slowing the synthesis of cholesterol. Degradation of HMGCR is inhibited by its binding to UBIAD1 (UbiA prenyltransferase domain-containing protein-1). This inhibition contributes to statin-induced accumulation of HMGCR, which limits their cholesterol-lowering effects. Here, we report cryo-electron microscopy structures of the HMGCR-UBIAD1 complex, which is maintained by interactions between transmembrane helix (TM) 7 of HMGCR and TMs 2-4 of UBIAD1. Disrupting this interface by mutagenesis prevents complex formation, enhancing HMGCR degradation. TMs 2-6 of HMGCR contain a 170-amino acid sterol sensing domain (SSD), which exists in two conformations-one of which is essential for degradation. Thus, our data supports a model that rearrangement of the TMs in the SSD permits recruitment of proteins that initate HMGCR degradation, a key reaction in the regulatory system that governs cholesterol synthesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27475.map.gz emd_27475.map.gz | 116.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27475-v30.xml emd-27475-v30.xml emd-27475.xml emd-27475.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27475.png emd_27475.png | 77.6 KB | ||
| Others |  emd_27475_half_map_1.map.gz emd_27475_half_map_1.map.gz emd_27475_half_map_2.map.gz emd_27475_half_map_2.map.gz | 98.5 MB 98.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27475 http://ftp.pdbj.org/pub/emdb/structures/EMD-27475 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27475 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27475 | HTTPS FTP |
-Validation report
| Summary document |  emd_27475_validation.pdf.gz emd_27475_validation.pdf.gz | 852.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27475_full_validation.pdf.gz emd_27475_full_validation.pdf.gz | 852.4 KB | Display | |
| Data in XML |  emd_27475_validation.xml.gz emd_27475_validation.xml.gz | 13.6 KB | Display | |
| Data in CIF |  emd_27475_validation.cif.gz emd_27475_validation.cif.gz | 16.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27475 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27475 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27475 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27475 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27475.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27475.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.842 Å | ||||||||||||||||||||||||||||||||||||
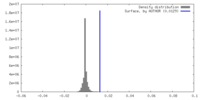
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27475_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
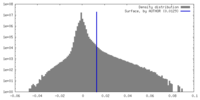
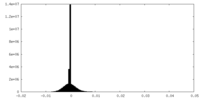
| Density Histograms |
-Half map: #2
| File | emd_27475_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
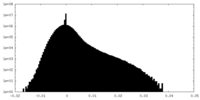
| Density Histograms |
- Sample components
Sample components
-Entire : HMGCR-UBIAD1-BRIL-Fab-Nb Complex
| Entire | Name: HMGCR-UBIAD1-BRIL-Fab-Nb Complex |
|---|---|
| Components |
|
-Supramolecule #1: HMGCR-UBIAD1-BRIL-Fab-Nb Complex
| Supramolecule | Name: HMGCR-UBIAD1-BRIL-Fab-Nb Complex / type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 8.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 258143 |
|---|---|
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)