[English] 日本語
 Yorodumi
Yorodumi- EMDB-26163: Nipah Virus attachment (G) glycoprotein ectodomain in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-26163 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nipah Virus attachment (G) glycoprotein ectodomain in complex with nAH1.3 neutralizing antibody Fab fragment (local refinement of the stalk region) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NiV / NiVG / attachment protein / neutralizing antibodies / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion involved in viral entry into host cell / exo-alpha-sialidase activity / immunoglobulin complex / clathrin-dependent endocytosis of virus by host cell / adaptive immune response / host cell surface receptor binding / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane ...membrane fusion involved in viral entry into host cell / exo-alpha-sialidase activity / immunoglobulin complex / clathrin-dependent endocytosis of virus by host cell / adaptive immune response / host cell surface receptor binding / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / metal ion binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Nipah henipavirus / Nipah henipavirus /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Wang ZQ / Seattle Structural Genomics Center for Infectious Disease (SSGCID) | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Architecture and antigenicity of the Nipah virus attachment glycoprotein. Authors: Zhaoqian Wang / Moushimi Amaya / Amin Addetia / Ha V Dang / Gabriella Reggiano / Lianying Yan / Andrew C Hickey / Frank DiMaio / Christopher C Broder / David Veesler /  Abstract: Nipah virus (NiV) and Hendra virus (HeV) are zoonotic henipaviruses (HNVs) responsible for outbreaks of encephalitis and respiratory illness. The entry of HNVs into host cells requires the attachment ...Nipah virus (NiV) and Hendra virus (HeV) are zoonotic henipaviruses (HNVs) responsible for outbreaks of encephalitis and respiratory illness. The entry of HNVs into host cells requires the attachment (G) and fusion (F) glycoproteins, which are the main targets of antibody responses. To understand viral infection and host immunity, we determined a cryo-electron microscopy structure of the NiV G homotetrameric ectodomain in complex with the nAH1.3 broadly neutralizing antibody Fab fragment. We show that a cocktail of two nonoverlapping G-specific antibodies neutralizes NiV and HeV synergistically and limits the emergence of escape mutants. Analysis of polyclonal serum antibody responses elicited by vaccination of macaques with NiV G indicates that the receptor binding head domain is immunodominant. These results pave the way for implementing multipronged therapeutic strategies against these deadly pathogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26163.map.gz emd_26163.map.gz | 350.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26163-v30.xml emd-26163-v30.xml emd-26163.xml emd-26163.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26163.png emd_26163.png | 73.4 KB | ||
| Filedesc metadata |  emd-26163.cif.gz emd-26163.cif.gz | 6.4 KB | ||
| Others |  emd_26163_additional_1.map.gz emd_26163_additional_1.map.gz emd_26163_half_map_1.map.gz emd_26163_half_map_1.map.gz emd_26163_half_map_2.map.gz emd_26163_half_map_2.map.gz | 186.6 MB 344.5 MB 344.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26163 http://ftp.pdbj.org/pub/emdb/structures/EMD-26163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26163 | HTTPS FTP |
-Related structure data
| Related structure data |  7ty0MC  7txzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26163.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26163.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_26163_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
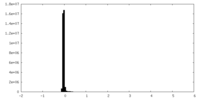
| Density Histograms |
-Half map: #2
| File | emd_26163_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
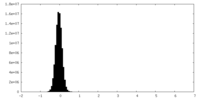
| Density Histograms |
-Half map: #1
| File | emd_26163_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
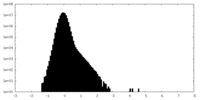
| Density Histograms |
- Sample components
Sample components
-Entire : Nipah Virus attachment protein ectodomain in complex with nAH1.3 ...
| Entire | Name: Nipah Virus attachment protein ectodomain in complex with nAH1.3 neutralizing antibody Fab fragment |
|---|---|
| Components |
|
-Supramolecule #1: Nipah Virus attachment protein ectodomain in complex with nAH1.3 ...
| Supramolecule | Name: Nipah Virus attachment protein ectodomain in complex with nAH1.3 neutralizing antibody Fab fragment type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Nipah henipavirus Nipah henipavirus |
-Macromolecule #1: Glycoprotein G
| Macromolecule | Name: Glycoprotein G / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nipah henipavirus Nipah henipavirus |
| Molecular weight | Theoretical: 60.358535 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: HHHHHHIQNY TRSTDNQAVI KDALQGIQQQ IKGLADKIGT EIGPKVSLID TSSTITIPAN IGLLGSKISQ STASINENVN EKCKFTLPP LKIHECNISC PNPLPFREYR PQTEGVSNLV GLPNNICLQK TSNQILKPKL ISYTLPVVGQ SGTCITDPLL A MDEGYFAY ...String: HHHHHHIQNY TRSTDNQAVI KDALQGIQQQ IKGLADKIGT EIGPKVSLID TSSTITIPAN IGLLGSKISQ STASINENVN EKCKFTLPP LKIHECNISC PNPLPFREYR PQTEGVSNLV GLPNNICLQK TSNQILKPKL ISYTLPVVGQ SGTCITDPLL A MDEGYFAY SHLERIGSCS RGVSKQRIIG VGEVLDRGDE VPSLFMTNVW TPPNPNTVYH CSAVYNNEFY YVLCAVSTVG DP ILNSTYW SGSLMMTRLA VKPKSNGGGY NQHQLALRSI EKGRYDKVMP YGPSGIKQGD TLYFPAVGFL VRTEFKYNDS NCP ITKCQY SKPENCRLSM GIRPNSHYIL RSGLLKYNLS DGENPKVVFI EISDQRLSIG SPSKIYDSLG QPVFYQASFS WDTM IKFGD VLTVNPLVVN WRNNTVISRP GQSQCPRFNT CPEICWEGVY NDAFLIDRIN WISAGVFLDS NQTAENPVFT VFKDN EILY RAQLASEDTN AQKTITNCFL LKNKIWCISL VEIYDTGDNV IRPKLFAVKI PEQCY UniProtKB: Glycoprotein G |
-Macromolecule #2: Igh protein
| Macromolecule | Name: Igh protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.643941 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVKLEESGGG LVQPGGSMKL SCVASGFSFS YYWMNWVRQS PEKGLEWVAE IRLKSNNYGT HYAESVKGRF TISRDDSKSS VYLQMNNLR PEDTGIYYCT RVITTVFAYW GQGTLVTVSA AKTTPPSVYP LAPGSAAQTN SMVTLGCLVK GYFPEPVTVT W NSGSLSSG ...String: EVKLEESGGG LVQPGGSMKL SCVASGFSFS YYWMNWVRQS PEKGLEWVAE IRLKSNNYGT HYAESVKGRF TISRDDSKSS VYLQMNNLR PEDTGIYYCT RVITTVFAYW GQGTLVTVSA AKTTPPSVYP LAPGSAAQTN SMVTLGCLVK GYFPEPVTVT W NSGSLSSG VHTFPAVLQS DLYTLSSSVT VPSSTWPSET VTCNVAHPAS STKVDKKIVP RDCGCKPCIC TVPEVSSVFI FP PKPKDVL TITLTPKVTC VVVDISKDDP EVQFSWFVDD VEVHTAQTQP REEQFNSTFR SVSELPIMHQ DWLNGKEFKC RVN SAAFPA PIEKTISKTK GRPKAPQVYT IPPPKEQMAK DKVSLTCMIT DFFPEDITVE WQWNGQPAEN YKNTQPIMDT DGSY FVYSK LNVQKSNWEA GNTFTCSVLH EGLHNHHTEK SLSHSPGKGG SGGGSWSHPQ FEK UniProtKB: Igh protein |
-Macromolecule #3: nAH Fab light chain
| Macromolecule | Name: nAH Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.947473 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVLTQSPAS LAVSLGQRAT ISCRASESVH DYGISFMNWF QQKPGQPPKL LIYSASNQGS GVPARFSGSG SGTDFSLNIH PMEEDDIAM YFCQQSKEVP YTFGGGTKLE IKRADAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT ...String: DIVLTQSPAS LAVSLGQRAT ISCRASESVH DYGISFMNWF QQKPGQPPKL LIYSASNQGS GVPARFSGSG SGTDFSLNIH PMEEDDIAM YFCQQSKEVP YTFGGGTKLE IKRADAAPTV SIFPPSSEQL TSGGASVVCF LNNFYPKDIN VKWKIDGSER Q NGVLNSWT DQDSKDSTYS MSSTLTLTKD EYERHNSYTC EATHKTSTSP IVKSFNRNEC |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 168035 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)