[English] 日本語
 Yorodumi
Yorodumi- EMDB-2556: Negative-stain electron microscopy of E. coli ClpB mutant E432A (... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2556 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP) | |||||||||
 Map data Map data | Asymmetric reconstruction of ClpB E432A mutant (BAP form bound to CLpP) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | chaperone / disaggregase / ClpB / BAP / repressed E432A mutant / coiled-coil domain | |||||||||
| Biological species |  | |||||||||
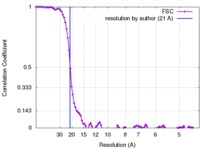
| Method | single particle reconstruction / negative staining / Resolution: 21.0 Å | |||||||||
 Authors Authors | Carroni M / Kummer E / Oguchi Y / Wendler P / Sinning I / Kopp J / Mogk A / Bukau B / Saibil HR | |||||||||
 Citation Citation |  Journal: Elife / Year: 2014 Journal: Elife / Year: 2014Title: Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Authors: Marta Carroni / Eva Kummer / Yuki Oguchi / Petra Wendler / Daniel K Clare / Irmgard Sinning / Jürgen Kopp / Axel Mogk / Bernd Bukau / Helen R Saibil /   Abstract: The hexameric AAA+ chaperone ClpB reactivates aggregated proteins in cooperation with the Hsp70 system. Essential for disaggregation, the ClpB middle domain (MD) is a coiled-coil propeller that binds ...The hexameric AAA+ chaperone ClpB reactivates aggregated proteins in cooperation with the Hsp70 system. Essential for disaggregation, the ClpB middle domain (MD) is a coiled-coil propeller that binds Hsp70. Although the ClpB subunit structure is known, positioning of the MD in the hexamer and its mechanism of action are unclear. We obtained electron microscopy (EM) structures of the BAP variant of ClpB that binds the protease ClpP, clearly revealing MD density on the surface of the ClpB ring. Mutant analysis and asymmetric reconstructions show that MDs adopt diverse positions in a single ClpB hexamer. Adjacent, horizontally oriented MDs form head-to-tail contacts and repress ClpB activity by preventing Hsp70 interaction. Tilting of the MD breaks this contact, allowing Hsp70 binding, and releasing the contact in adjacent subunits. Our data suggest a wavelike activation of ClpB subunits around the ring.DOI: http://dx.doi.org/10.7554/eLife.02481.001. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2556.map.gz emd_2556.map.gz | 61.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2556-v30.xml emd-2556-v30.xml emd-2556.xml emd-2556.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2556_fsc.xml emd_2556_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_2556.tiff emd_2556.tiff | 352.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2556 http://ftp.pdbj.org/pub/emdb/structures/EMD-2556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2556 | HTTPS FTP |
-Validation report
| Summary document |  emd_2556_validation.pdf.gz emd_2556_validation.pdf.gz | 214.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2556_full_validation.pdf.gz emd_2556_full_validation.pdf.gz | 213.8 KB | Display | |
| Data in XML |  emd_2556_validation.xml.gz emd_2556_validation.xml.gz | 10.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2556 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2556 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2556 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2556 | HTTPS FTP |
-Related structure data
| Related structure data |  2555C  2557C  2558C 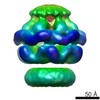 2559C 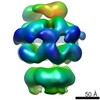 2560C  2561C  2562C  2563C  4ciuC  4d2qC  4d2uC  4d2xC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2556.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2556.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetric reconstruction of ClpB E432A mutant (BAP form bound to CLpP) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ClpB E432A ATPgammaS. BAP variant bound to ClpP.
| Entire | Name: ClpB E432A ATPgammaS. BAP variant bound to ClpP. |
|---|---|
| Components |
|
-Supramolecule #1000: ClpB E432A ATPgammaS. BAP variant bound to ClpP.
| Supramolecule | Name: ClpB E432A ATPgammaS. BAP variant bound to ClpP. / type: sample / ID: 1000 Details: Only the ClpB part was reconstructed and the molecular weight only refers to this part. Oligomeric state: Homohexamer. (One homohexamer of BAP bound to one homoheptamer of ClpP) Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: ClpB
| Macromolecule | Name: ClpB / type: protein_or_peptide / ID: 1 / Details: The protein is engineered to bind to ClpP. / Number of copies: 6 / Oligomeric state: Hexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 80 KDa / Theoretical: 80 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM Tris-HCl, pH 7.5, 20 mM KCl, 15 mM MgCl2, 1 mM DTT, 2 mM ATPgammaS |
| Staining | Type: NEGATIVE Details: protein adsorbed on carbon coated grids pretreated with 0.01% poly lysine chains. Stained with 2% uranyl acetate for 1 minute. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 150,000 x magnification |
| Date | Jun 6, 2011 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 110 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 68000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Fitting of separate domains was performed manually and locally optimised using Chimera. Known domain interfaces were used to guide the fit. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Fitting of separate domains was performed manually and locally optimised using Chimera. Known domain interfaces were used to guide the fit. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)