[English] 日本語
 Yorodumi
Yorodumi- EMDB-25437: Structure of DPSL (DNA Protection in Starved Cells - Like) from P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25437 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
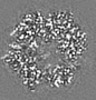
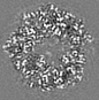
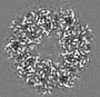
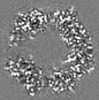
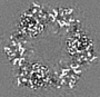
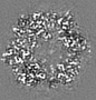
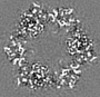
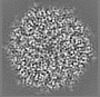
| Title | Structure of DPSL (DNA Protection in Starved Cells - Like) from Pyrococcus furiosus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DPS-like protein / METAL BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationOxidoreductases; Oxidizing metal ions / nucleoid / ferric iron binding / intracellular iron ion homeostasis / oxidoreductase activity / DNA binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.37 Å | |||||||||
 Authors Authors | Gauvin CC / Waghwani HK | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: Structure of DPSL (DNA Protection in Starved Cells - Like) from Pyrococcus furiosus Authors: Ramsay B / Wiedenheft B / Allen M / Gauss GH / Lawrence CM / Young M / Douglas T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25437.map.gz emd_25437.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25437-v30.xml emd-25437-v30.xml emd-25437.xml emd-25437.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25437_fsc.xml emd_25437_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_25437.png emd_25437.png | 142.9 KB | ||
| Filedesc metadata |  emd-25437.cif.gz emd-25437.cif.gz | 6.4 KB | ||
| Others |  emd_25437_half_map_1.map.gz emd_25437_half_map_1.map.gz emd_25437_half_map_2.map.gz emd_25437_half_map_2.map.gz | 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25437 http://ftp.pdbj.org/pub/emdb/structures/EMD-25437 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25437 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25437 | HTTPS FTP |
-Validation report
| Summary document |  emd_25437_validation.pdf.gz emd_25437_validation.pdf.gz | 965 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25437_full_validation.pdf.gz emd_25437_full_validation.pdf.gz | 964.6 KB | Display | |
| Data in XML |  emd_25437_validation.xml.gz emd_25437_validation.xml.gz | 10.6 KB | Display | |
| Data in CIF |  emd_25437_validation.cif.gz emd_25437_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25437 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25437 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25437 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25437 | HTTPS FTP |
-Related structure data
| Related structure data |  7stwMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25437.map.gz / Format: CCP4 / Size: 2.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25437.map.gz / Format: CCP4 / Size: 2.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.152 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1
| File | emd_25437_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
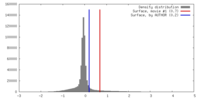
| Density Histograms |
-Half map: Half map 2
| File | emd_25437_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
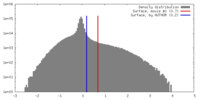
| Density Histograms |
- Sample components
Sample components
-Entire : Dodecameric assembly of DPS-like protein
| Entire | Name: Dodecameric assembly of DPS-like protein |
|---|---|
| Components |
|
-Supramolecule #1: Dodecameric assembly of DPS-like protein
| Supramolecule | Name: Dodecameric assembly of DPS-like protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Theoretical: 256.32 KDa |
-Macromolecule #1: DNA protection during starvation protein
| Macromolecule | Name: DNA protection during starvation protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number: Oxidoreductases; Oxidizing metal ions |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) / Strain: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 Pyrococcus furiosus (archaea) / Strain: ATCC 43587 / DSM 3638 / JCM 8422 / Vc1 |
| Molecular weight | Theoretical: 21.387254 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPEHNRRLVE RTGIDVEKLL ELLIKAAAAE FTTYYYYTIL RNHATGLEGE AIKEIIEDAR LEDRNHFEAL VPRIYELGGE LPRDIREFA DLASCRDAYL PEEPTIENIL KVLLEAERCA VGVYTEICNY TFGKDPRTYD LALAILHEEI EHEAWFEELL T GKPSGHFR RGKPGESPYV SKFLKTR UniProtKB: DNA protection during starvation protein |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 12 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 3 / Number of copies: 12 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 257 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force 5 Blot time 4. |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 100.0 K |
| Details | Beam tilt was performed with a 3x3 matrix with corners omitted. |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 1574 / Average exposure time: 7.0 sec. / Average electron dose: 65.0 e/Å2 / Details: Images were dose-fractionated to have 50 frames |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.2 µm / Calibrated defocus min: 0.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.62 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Workflow involved generating a homology model from 2CLB structure from S. solfataricus with the Pf sequence, using Swiss Model. Then, that homology model was fit to the map density initially in Chimera using fitmap. After that, it was initially refined with rigid body real-space refinement in Phenix, tuned in coot, and subsequently finished refining with flexible fitting and enforced tetrahedral symmetry. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-7stw: |
 Movie
Movie Controller
Controller














 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)