[English] 日本語
 Yorodumi
Yorodumi- EMDB-25361: Composite map of ciliary C2 central pair apparatus isolated from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Composite map of ciliary C2 central pair apparatus isolated from Chlamydomonas reinhardtii | |||||||||
 Map data Map data | Ciliary C2 central pair apparatus isolated from Chlamydomonas reinhardtii | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cilia / microtubule / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationaxonemal central pair / axonemal doublet microtubule / organelle / positive regulation of cilium-dependent cell motility / regulation of cilium beat frequency involved in ciliary motility / establishment of protein localization to organelle / cilium movement / axoneme assembly / axonemal microtubule / motile cilium ...axonemal central pair / axonemal doublet microtubule / organelle / positive regulation of cilium-dependent cell motility / regulation of cilium beat frequency involved in ciliary motility / establishment of protein localization to organelle / cilium movement / axoneme assembly / axonemal microtubule / motile cilium / microtubule associated complex / regulation of cytoskeleton organization / axoneme / microtubule-based process / cilium assembly / ciliary basal body / structural constituent of cytoskeleton / microtubule binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / transcription coactivator activity / hydrolase activity / GTPase activity / calcium ion binding / regulation of DNA-templated transcription / GTP binding / nucleus / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
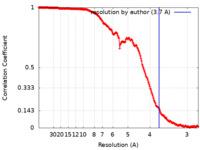
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Gui M / Wang X / Dutcher SK / Brown A / Zhang R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Ciliary central apparatus structure reveals mechanisms of microtubule patterning. Authors: Miao Gui / Xiangli Wang / Susan K Dutcher / Alan Brown / Rui Zhang /  Abstract: A pair of extensively modified microtubules form the central apparatus (CA) of the axoneme of most motile cilia, where they regulate ciliary motility. The external surfaces of both CA microtubules ...A pair of extensively modified microtubules form the central apparatus (CA) of the axoneme of most motile cilia, where they regulate ciliary motility. The external surfaces of both CA microtubules are patterned asymmetrically with large protein complexes that repeat every 16 or 32 nm. The composition of these projections and the mechanisms that establish asymmetry and longitudinal periodicity are unknown. Here, by determining cryo-EM structures of the CA microtubules, we identify 48 different CA-associated proteins, which in turn reveal mechanisms for asymmetric and periodic protein binding to microtubules. We identify arc-MIPs, a novel class of microtubule inner protein, that bind laterally across protofilaments and remodel tubulin structure and lattice contacts. The binding mechanisms utilized by CA proteins may be generalizable to other microtubule-associated proteins. These structures establish a foundation to elucidate the contributions of individual CA proteins to ciliary motility and ciliopathies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25361.map.gz emd_25361.map.gz | 246.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25361-v30.xml emd-25361-v30.xml emd-25361.xml emd-25361.xml | 52.5 KB 52.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25361_fsc.xml emd_25361_fsc.xml | 17.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_25361.png emd_25361.png | 76 KB | ||
| Masks |  emd_25361_msk_1.map emd_25361_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25361.cif.gz emd-25361.cif.gz | 11.9 KB | ||
| Others |  emd_25361_additional_1.map.gz emd_25361_additional_1.map.gz emd_25361_additional_2.map.gz emd_25361_additional_2.map.gz emd_25361_additional_3.map.gz emd_25361_additional_3.map.gz emd_25361_additional_4.map.gz emd_25361_additional_4.map.gz emd_25361_additional_5.map.gz emd_25361_additional_5.map.gz emd_25361_additional_6.map.gz emd_25361_additional_6.map.gz emd_25361_additional_7.map.gz emd_25361_additional_7.map.gz emd_25361_additional_8.map.gz emd_25361_additional_8.map.gz emd_25361_additional_9.map.gz emd_25361_additional_9.map.gz | 1.9 MB 1.9 MB 1.9 MB 1.9 MB 257.5 MB 4.4 MB 1.9 MB 1.9 MB 1.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25361 http://ftp.pdbj.org/pub/emdb/structures/EMD-25361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25361 | HTTPS FTP |
-Validation report
| Summary document |  emd_25361_validation.pdf.gz emd_25361_validation.pdf.gz | 557.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25361_full_validation.pdf.gz emd_25361_full_validation.pdf.gz | 557.3 KB | Display | |
| Data in XML |  emd_25361_validation.xml.gz emd_25361_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_25361_validation.cif.gz emd_25361_validation.cif.gz | 22.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25361 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25361 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25361 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25361 | HTTPS FTP |
-Related structure data
| Related structure data |  7somMC  7sqcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25361.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25361.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ciliary C2 central pair apparatus isolated from Chlamydomonas reinhardtii | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Mask #1
+Additional map: Additional map 1
+Additional map: Additional map 2
+Additional map: Additional map 3
+Additional map: Additional map 4
+Additional map: consensus refinement
+Additional map: mask for fsc curve
+Additional map: Additional map 7
+Additional map: Additional map 8
+Additional map: Additional map 9
- Sample components
Sample components
+Entire : C2 central pair apparatus complex
+Supramolecule #1: C2 central pair apparatus complex
+Macromolecule #1: Tubulin beta
+Macromolecule #2: Tubulin alpha
+Macromolecule #3: Cilia- and flagella-associated protein 20
+Macromolecule #4: Unknown protein
+Macromolecule #5: Unknown protein
+Macromolecule #6: Unknown protein
+Macromolecule #7: FAP65
+Macromolecule #8: FAP70
+Macromolecule #9: FAP147
+Macromolecule #10: FAP178
+Macromolecule #11: Flagellar WD repeat-containing protein Pf20
+Macromolecule #12: Flagellar associated protein
+Macromolecule #13: FAP196
+Macromolecule #14: FAP213
+Macromolecule #15: FAP225
+Macromolecule #16: FAP239
+Macromolecule #17: FAP388
+Macromolecule #18: FAP424
+Macromolecule #19: GUANOSINE-5'-DIPHOSPHATE
+Macromolecule #20: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #21: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 39.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)