+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23542 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of full-length GluK1 with L-Glu | |||||||||
 Map data Map data | GluK1 with L-Glu (full-length | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / glutamate receptor / kainate receptor / MEMBRANE PROTEIN | |||||||||
| Function / homology | Isoform Glur5-2 of Glutamate receptor ionotropic, kainate 1 Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Meyerson JR / Selvakumar P | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Structural and compositional diversity in the kainate receptor family. Authors: Purushotham Selvakumar / Joon Lee / Nandish Khanra / Changhao He / Hermany Munguba / Lisa Kiese / Johannes Broichhagen / Andreas Reiner / Joshua Levitz / Joel R Meyerson /   Abstract: The kainate receptors (KARs) are members of the ionotropic glutamate receptor family and assemble into tetramers from a pool of five subunit types (GluK1-5). Each subunit confers distinct functional ...The kainate receptors (KARs) are members of the ionotropic glutamate receptor family and assemble into tetramers from a pool of five subunit types (GluK1-5). Each subunit confers distinct functional properties to a receptor, but the compositional and stoichiometric diversity of KAR tetramers is not well understood. To address this, we first solve the structure of the GluK1 homomer, which enables a systematic assessment of structural compatibility among KAR subunits. Next, we analyze single-cell RNA sequencing data, which reveal extreme diversity in the combinations of two or more KAR subunits co-expressed within the same cell. We then investigate the composition of individual receptor complexes using single-molecule fluorescence techniques and find that di-heteromers assembled from GluK1, GluK2, or GluK3 can form with all possible stoichiometries, while GluK1/K5, GluK2/K5, and GluK3/K5 can form 3:1 or 2:2 complexes. Finally, using three-color single-molecule imaging, we discover that KARs can form tri- and tetra-heteromers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23542.map.gz emd_23542.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23542-v30.xml emd-23542-v30.xml emd-23542.xml emd-23542.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23542_fsc.xml emd_23542_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_23542.png emd_23542.png | 54 KB | ||
| Filedesc metadata |  emd-23542.cif.gz emd-23542.cif.gz | 5.4 KB | ||
| Others |  emd_23542_additional_1.map.gz emd_23542_additional_1.map.gz emd_23542_additional_2.map.gz emd_23542_additional_2.map.gz | 2.3 MB 98.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23542 http://ftp.pdbj.org/pub/emdb/structures/EMD-23542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23542 | HTTPS FTP |
-Validation report
| Summary document |  emd_23542_validation.pdf.gz emd_23542_validation.pdf.gz | 556 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23542_full_validation.pdf.gz emd_23542_full_validation.pdf.gz | 555.6 KB | Display | |
| Data in XML |  emd_23542_validation.xml.gz emd_23542_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_23542_validation.cif.gz emd_23542_validation.cif.gz | 16.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23542 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23542 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23542 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23542 | HTTPS FTP |
-Related structure data
| Related structure data |  7lvtMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23542.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23542.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK1 with L-Glu (full-length | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.096 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
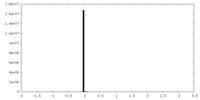
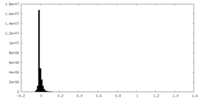
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: GluK1 with L-Glu (ATD)
| File | emd_23542_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK1 with L-Glu (ATD) | ||||||||||||
| Projections & Slices |
| ||||||||||||
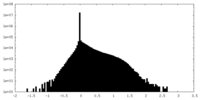
| Density Histograms |
-Additional map: GluK1 with L-Glu (LBD-TMD)
| File | emd_23542_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GluK1 with L-Glu (LBD-TMD) | ||||||||||||
| Projections & Slices |
| ||||||||||||
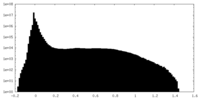
| Density Histograms |
- Sample components
Sample components
-Entire : GluK1 tetrameric complex
| Entire | Name: GluK1 tetrameric complex |
|---|---|
| Components |
|
-Supramolecule #1: GluK1 tetrameric complex
| Supramolecule | Name: GluK1 tetrameric complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Isoform Glur5-2 of Glutamate receptor ionotropic, kainate 1
| Macromolecule | Name: Isoform Glur5-2 of Glutamate receptor ionotropic, kainate 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 102.562078 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKIISPVLSN LVFSRSIKVL LCLLWIGYSQ GTTHVLRIGG IFETVENEPV NVEELAFKFA VTSINRNRTL MPNTTLTYDI QRINLFDSF EASRRACDQL ALGVAALFGP SHSSSVSAVQ SICNALEVPH IQTRWKHPSV DSRDLFYINL YPDYAAISRA V LDLVLYYN ...String: MKIISPVLSN LVFSRSIKVL LCLLWIGYSQ GTTHVLRIGG IFETVENEPV NVEELAFKFA VTSINRNRTL MPNTTLTYDI QRINLFDSF EASRRACDQL ALGVAALFGP SHSSSVSAVQ SICNALEVPH IQTRWKHPSV DSRDLFYINL YPDYAAISRA V LDLVLYYN WKTVTVVYED STGLIRLQEL IKAPSRYNIK IKIRQLPPAN KDAKPLLKEM KKSKEFYVIF DCSHETAAEI LK QILFMGM MTEYYHYFFT TLDLFALDLE LYRYSGVNMT GFRLLNIDNP HVSSIIEKWS MERLQAPPRP ETGLLDGMMT TEA ALMYDA VYMVAIASHR ASQLTVSSLQ CHRHKPWRLG PRFMNLIKEA RWDGLTGRIT FNKTDGLRKD FDLDIISLKE EGTE KIGIW NSNSGLNMTD GNRDRSNNIT DSLANRTLIV TTILEEPYVM YRKSDKPLYG NDRFEGYCLD LLKELSNILG FLYDV KLVP DGKYGAQNDK GEWNGMVKEL IDHRADLAVA PLTITYVREK VIDFSKPFMT LGISILYRKP NGTNPGVFSF LNPLSP DIW MYVLLACLGV SCVLFVIARF TPYEWYNPHP CNPDSDVVEN NFTLLNSFWF GVGALMQQGS ELMPKALSTR IVGGIWW FF TLIIISSYTA NLAAFLTVER MESPIDSADD LAKQTKIEYG AVRDGSTMTF FKKSKISTYE KMWAFMSSRQ QSALVKNS D EGIQRVLTTD YALLMESTSI EYVTQRNCNL TQIGGLIDSK GYGVGTPIGS PYRDKITIAI LQLQEEGKLH MMKEKWWRG NGCPEEDSKE ASALGVENIG GIFIVLAAGL VLSVFVAIGE FLYKSRKNND VEQCLSFNAI MEELGISLKN QKKLKKKSRT KGKSSFTSI LTCHQRRTQR KETVA UniProtKB: Isoform Glur5-2 of Glutamate receptor ionotropic, kainate 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)