[English] 日本語
 Yorodumi
Yorodumi- EMDB-23236: BG505 SOSIP.v5.2 N241/N289 in complex with the polyclonal Fab pAb... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23236 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BG505 SOSIP.v5.2 N241/N289 in complex with the polyclonal Fab pAbC-1 from animal Rh.33311 (Wk26 time point) | |||||||||
 Map data Map data | 3D map of BG505 SOSIP v5.2 N241/N289 in complex with the polyclonal Fab pAbC-1 from animal Rh.33311 (Wk26 time point). Main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV / vaccine design / BG505 / VIRAL PROTEIN-IMMUNE SYSTEM complex / Polyclonal antibodies / EMPEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Antanasijevic A / Sewall LM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM. Authors: Aleksandar Antanasijevic / Leigh M Sewall / Christopher A Cottrell / Diane G Carnathan / Luis E Jimenez / Julia T Ngo / Jennifer B Silverman / Bettina Groschel / Erik Georgeson / Jinal ...Authors: Aleksandar Antanasijevic / Leigh M Sewall / Christopher A Cottrell / Diane G Carnathan / Luis E Jimenez / Julia T Ngo / Jennifer B Silverman / Bettina Groschel / Erik Georgeson / Jinal Bhiman / Raiza Bastidas / Celia LaBranche / Joel D Allen / Jeffrey Copps / Hailee R Perrett / Kimmo Rantalainen / Fabien Cannac / Yuhe R Yang / Alba Torrents de la Peña / Rebeca Froes Rocha / Zachary T Berndsen / David Baker / Neil P King / Rogier W Sanders / John P Moore / Shane Crotty / Max Crispin / David C Montefiori / Dennis R Burton / William R Schief / Guido Silvestri / Andrew B Ward /    Abstract: Engineered ectodomain trimer immunogens based on BG505 envelope glycoprotein are widely utilized as components of HIV vaccine development platforms. In this study, we used rhesus macaques to evaluate ...Engineered ectodomain trimer immunogens based on BG505 envelope glycoprotein are widely utilized as components of HIV vaccine development platforms. In this study, we used rhesus macaques to evaluate the immunogenicity of several stabilized BG505 SOSIP constructs both as free trimers and presented on a nanoparticle. We applied a cryoEM-based method for high-resolution mapping of polyclonal antibody responses elicited in immunized animals (cryoEMPEM). Mutational analysis coupled with neutralization assays were used to probe the neutralization potential at each epitope. We demonstrate that cryoEMPEM data can be used for rapid, high-resolution analysis of polyclonal antibody responses without the need for monoclonal antibody isolation. This approach allowed to resolve structurally distinct classes of antibodies that bind overlapping sites. In addition to comprehensive mapping of commonly targeted neutralizing and non-neutralizing epitopes in BG505 SOSIP immunogens, our analysis revealed that epitopes comprising engineered stabilizing mutations and of partially occupied glycosylation sites can be immunogenic. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23236.map.gz emd_23236.map.gz | 165.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23236-v30.xml emd-23236-v30.xml emd-23236.xml emd-23236.xml | 27.5 KB 27.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23236_fsc.xml emd_23236_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_23236.png emd_23236.png | 163.8 KB | ||
| Masks |  emd_23236_msk_1.map emd_23236_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23236.cif.gz emd-23236.cif.gz | 7.8 KB | ||
| Others |  emd_23236_half_map_1.map.gz emd_23236_half_map_1.map.gz emd_23236_half_map_2.map.gz emd_23236_half_map_2.map.gz | 140.5 MB 140.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23236 http://ftp.pdbj.org/pub/emdb/structures/EMD-23236 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23236 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23236 | HTTPS FTP |
-Validation report
| Summary document |  emd_23236_validation.pdf.gz emd_23236_validation.pdf.gz | 1021.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23236_full_validation.pdf.gz emd_23236_full_validation.pdf.gz | 1020.8 KB | Display | |
| Data in XML |  emd_23236_validation.xml.gz emd_23236_validation.xml.gz | 20 KB | Display | |
| Data in CIF |  emd_23236_validation.cif.gz emd_23236_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23236 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23236 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23236 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23236 | HTTPS FTP |
-Related structure data
| Related structure data |  7l8tMC  7l7tC  7l7uC  7l85C  7l86C  7l87C  7l88C  7l89C  7l8aC  7l8bC  7l8cC  7l8dC  7l8eC  7l8fC  7l8gC  7l8sC  7l8uC  7l8wC  7l8xC  7l8yC  7l8zC  7l90C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23236.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23236.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of BG505 SOSIP v5.2 N241/N289 in complex with the polyclonal Fab pAbC-1 from animal Rh.33311 (Wk26 time point). Main map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23236_msk_1.map emd_23236_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 3D map of BG505 SOSIP v5.2 N241/N289 in...
| File | emd_23236_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of BG505 SOSIP v5.2 N241/N289 in complex with the polyclonal Fab pAbC-1 from animal Rh.33311 (Wk26 time point). Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 3D map of BG505 SOSIP v5.2 N241/N289 in...
| File | emd_23236_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of BG505 SOSIP v5.2 N241/N289 in complex with the polyclonal Fab pAbC-1 from animal Rh.33311 (Wk26 time point). Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BG505 SOSIP.v5.2 N241/N289 in complex with the polyclonal Fab pAb...
| Entire | Name: BG505 SOSIP.v5.2 N241/N289 in complex with the polyclonal Fab pAbC-1 from animal Rh.33311 (Wk26 time point) |
|---|---|
| Components |
|
-Supramolecule #1: BG505 SOSIP.v5.2 N241/N289 in complex with the polyclonal Fab pAb...
| Supramolecule | Name: BG505 SOSIP.v5.2 N241/N289 in complex with the polyclonal Fab pAbC-1 from animal Rh.33311 (Wk26 time point) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Polyclonal complexes were generated by incubation of isolated polyclonal Fabs with recombinantly expressed BG505 SOSIP and subsequent SEC purification. Map and model were reconstructed from ...Details: Polyclonal complexes were generated by incubation of isolated polyclonal Fabs with recombinantly expressed BG505 SOSIP and subsequent SEC purification. Map and model were reconstructed from the immune complex dataset using the focused classification approach. |
|---|
-Supramolecule #2: BG505 SOSIP.v5.2 N241/N289
| Supramolecule | Name: BG505 SOSIP.v5.2 N241/N289 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #3: polyclonal Fab pAbC-1 from animal Rh.33311
| Supramolecule | Name: polyclonal Fab pAbC-1 from animal Rh.33311 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: BG505 SOSIP.v5.2 N241/N289 - gp120
| Macromolecule | Name: BG505 SOSIP.v5.2 N241/N289 - gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 56.417125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKRGLCCVLL LCGAVFVSPS QEIHARFRRG ARAENLWVTV YYGVPVWKDA ETTLFCASDA KAYETKKHNV WATHCCVPTD PNPQEIHLE NVTEEFNMWK NNMVEQMHTD IISLWDQSLK PCVKLTPLCV TLQCTNVTNN ITDDMRGELK NCSFNMTTEL R DKKQKVYS ...String: MKRGLCCVLL LCGAVFVSPS QEIHARFRRG ARAENLWVTV YYGVPVWKDA ETTLFCASDA KAYETKKHNV WATHCCVPTD PNPQEIHLE NVTEEFNMWK NNMVEQMHTD IISLWDQSLK PCVKLTPLCV TLQCTNVTNN ITDDMRGELK NCSFNMTTEL R DKKQKVYS LFYRLDVVQI NENQGNRSNN SNKEYRLINC NTSAITQACP KVSFEPIPIH YCAPAGFAIL KCKDKKFNGT GP CTNVSTV QCTHGIKPVV STQLLLNGSL AEEEVIIRSE NITNNAKNIL VQLNESVQIN CTRPNNNTRK SIRIGPGQWF YAT GDIIGD IRQAHCNVSK ATWNETLGKV VKQLRKHFGN NTIIRFANSS GGDLEVTTHS FNCGGEFFYC NTSGLFNSTW ISNT SVQGS NSTGSNDSIT LPCRIKQIIN MWQRIGQAMY APPIQGVIRC VSNITGLILT RDGGSTNSTT ETFRPGGGDM RDNWR SELY KYKVVKIEPL GVAPTRCKR |
-Macromolecule #2: BG505 SOSIP.v5.2 N241/N289 - gp41
| Macromolecule | Name: BG505 SOSIP.v5.2 N241/N289 - gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.192574 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEC QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSTWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #3: Rh.33311 pAbC-1 - Heavy Chain
| Macromolecule | Name: Rh.33311 pAbC-1 - Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.890183 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK) |
-Macromolecule #4: Rh.33311 pAbC-1 - Light Chain
| Macromolecule | Name: Rh.33311 pAbC-1 - Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.698714 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 54 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: TBS buffer prepared from a 10X stock | |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time varied between 3 and 7 seconds.. | |||||||||
| Details | Polyclonal complexes were generated by incubation of isolated polyclonal Fabs with recombinantly expressed BG505 SOSIP and subsequent SEC purification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-39 / Number grids imaged: 4 / Number real images: 6268 / Average exposure time: 9.75 sec. / Average electron dose: 44.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||
| Output model |  PDB-7l8t: |
 Movie
Movie Controller
Controller









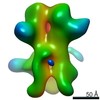
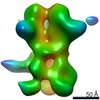

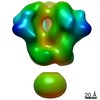































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)