[English] 日本語
 Yorodumi
Yorodumi- EMDB-8454: Cryo-electron microscopy structure of a bovine CLC-K chloride cha... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8454 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy structure of a bovine CLC-K chloride channel, auxiliary (class 2) conformation | |||||||||
 Map data Map data | Structure of a bovine CLC-K chloride channel. Combined, unfiltered map of Class 2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CLC / chloride channel / membrane / kidney / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationStimuli-sensing channels / voltage-gated chloride channel activity / chloride transport / chloride channel complex / basolateral plasma membrane / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
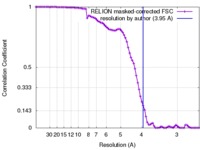
| Method | single particle reconstruction / cryo EM / Resolution: 3.95 Å | |||||||||
 Authors Authors | Park E / MacKinnon R | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Structure of a CLC chloride ion channel by cryo-electron microscopy. Authors: Eunyong Park / Ernest B Campbell / Roderick MacKinnon /  Abstract: CLC proteins transport chloride (Cl) ions across cellular membranes to regulate muscle excitability, electrolyte movement across epithelia, and acidification of intracellular organelles. Some CLC ...CLC proteins transport chloride (Cl) ions across cellular membranes to regulate muscle excitability, electrolyte movement across epithelia, and acidification of intracellular organelles. Some CLC proteins are channels that conduct Cl ions passively, whereas others are secondary active transporters that exchange two Cl ions for one H. The structural basis underlying these distinctive transport mechanisms is puzzling because CLC channels and transporters are expected to share the same architecture on the basis of sequence homology. Here we determined the structure of a bovine CLC channel (CLC-K) using cryo-electron microscopy. A conserved loop in the Cl transport pathway shows a structure markedly different from that of CLC transporters. Consequently, the cytosolic constriction for Cl passage is widened in CLC-K such that the kinetic barrier previously postulated for Cl/H transporter function would be reduced. Thus, reduction of a kinetic barrier in CLC channels enables fast flow of Cl down its electrochemical gradient. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8454.map.gz emd_8454.map.gz | 39.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8454-v30.xml emd-8454-v30.xml emd-8454.xml emd-8454.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8454_fsc.xml emd_8454_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_8454.png emd_8454.png | 59.6 KB | ||
| Masks |  emd_8454_msk_1.map emd_8454_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-8454.cif.gz emd-8454.cif.gz | 6.7 KB | ||
| Others |  emd_8454_additional_1.map.gz emd_8454_additional_1.map.gz emd_8454_additional_2.map.gz emd_8454_additional_2.map.gz | 49.3 MB 49.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8454 http://ftp.pdbj.org/pub/emdb/structures/EMD-8454 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8454 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8454 | HTTPS FTP |
-Related structure data
| Related structure data |  5tr1MC  8435C  5tqqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8454.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8454.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of a bovine CLC-K chloride channel. Combined, unfiltered map of Class 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_8454_msk_1.map emd_8454_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: B-factor-sharpend (-120 A^2) and lowpass-filtered (4.1 Ang)
| File | emd_8454_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor-sharpend (-120 A^2) and lowpass-filtered (4.1 Ang) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: B-factor-sharpend (-120 A^2) and lowpass-filtered (3.9 Ang)
| File | emd_8454_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor-sharpend (-120 A^2) and lowpass-filtered (3.9 Ang) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : A bovine CLC-K channel complexed with a monoclonal antibody fragm...
| Entire | Name: A bovine CLC-K channel complexed with a monoclonal antibody fragment (Fab) |
|---|---|
| Components |
|
-Supramolecule #1: A bovine CLC-K channel complexed with a monoclonal antibody fragm...
| Supramolecule | Name: A bovine CLC-K channel complexed with a monoclonal antibody fragment (Fab) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Fab fragment generated by proteolytic (papain) cleavage of murine IgG antibody. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Chloride channel protein
| Macromolecule | Name: Chloride channel protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 73.521484 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRVRRGIRGG LDWLKRKLFC VGEDWYFLTV LGVLMALISF TMSFTVGRVV RAHKWLYREI GDSHLLRYLS WTVYPVALVS FSSGFSQSI TPFSGGSGIP ELKTILSGVV LEDYLDIKNF GAKAVGLTCT LASGSTIFLG KVGPFVHLSV MIAAYLGRVR A KATGESEN ...String: MRVRRGIRGG LDWLKRKLFC VGEDWYFLTV LGVLMALISF TMSFTVGRVV RAHKWLYREI GDSHLLRYLS WTVYPVALVS FSSGFSQSI TPFSGGSGIP ELKTILSGVV LEDYLDIKNF GAKAVGLTCT LASGSTIFLG KVGPFVHLSV MIAAYLGRVR A KATGESEN KSKRNEMLVA GAAVGVATVF AAPFSGVLFC IEVVSSHFSV WDYWRGFFAA TCGAFMFRLL AVFNSEQETI TS LYKTSFR VEVPFDLPEI FFFVALGAIC GVASCAYLFC QRKFLGFVKT NPVLSKLMAT SKPLYSALAA LVLASVTYPP GAG RFMASR LSMREYLDSL LDHNSWALLT RQASPPWPVE PDPQNLWFEW YHPQFTIFGT LAFFLVMKFW MLILATTIPM PAGY FMPIF IFGAAIGRLL GEALSVAFPE GIVAGGVTNP IMPGGYALAG AAAFSGAVTH SISTALLAFE LTGQIVHALP VLMAV LAAN AIAQSCQPSF YDGTIIVKKL PYLPWIRGRK ISSHRVTVEH FMNRAITTLA KDTPQEEVVK VVTSTDMAEY PLVAST ESQ TLVGTMRRAQ LVQALQAEPP SWAPGQQRCL QDILAEGCPV EPVTLKLSPE TSLHQAHNLF ELLNLQSLFV TSQGRAV GF VSWVELEKAI SKLTNPPAPK SNSLEVLFQ UniProtKB: Chloride voltage-gated channel Ka |
-Macromolecule #2: Monoclonal antibody, Fab fragment, heavy chain
| Macromolecule | Name: Monoclonal antibody, Fab fragment, heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.633068 KDa |
| Sequence | String: DVQLQESGPG LVKPSQSLSL TCTVTGDSVT SDYAWSWIRQ FPGKKLEWMG YITYSGNTIY NPSLKSRISI TRDTSKNQFF LQLKSVIIE DTATYYCSRG VDYWGQGTSV TVSS |
-Macromolecule #3: Monoclonal antibody, Fab fragment, light chain
| Macromolecule | Name: Monoclonal antibody, Fab fragment, light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.824102 KDa |
| Sequence | String: DIVMTQSPKF MSTSVGDRVS VTCKASQNVG TNVAWYQQKP GQSPKTLIYW ASYRYSGVPD RFTGSGSGTD FTLAISNVQS EDLAEYFCQ QYNSYPLTFG SGTKLELK |
-Macromolecule #4: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 0.3 sec. / Average electron dose: 1.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)