[English] 日本語
 Yorodumi
Yorodumi- EMDB-22988: Adeno-associated virus serotype 5 in complex with the cellular re... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22988 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
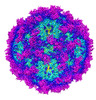
| Title | Adeno-associated virus serotype 5 in complex with the cellular receptor AAVR at 2.5 Angstroms resolution, AAV5 AAVR | |||||||||
 Map data Map data | Adeno-associated virus serotype 5 bound to its cellular receptor AAVR, cryo-em sharpened map at 2.5 Angstroms resolution. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAV5 / AAV / AAV-5 / AAVR / Adeno Associated Virus / PKD domain / VIRUS LIKE PARTICLE / parvovirus / virus / gene therapy / Receptor / Adeno-associated virus receptor / PKD1 / PKD | |||||||||
| Function / homology |  Function and homology information Function and homology informationproacrosomal vesicle fusion / response to auditory stimulus / flagellated sperm motility / T=1 icosahedral viral capsid / receptor-mediated endocytosis of virus by host cell / trans-Golgi network / neuron migration / cytoplasmic vesicle / Golgi membrane / nucleolus ...proacrosomal vesicle fusion / response to auditory stimulus / flagellated sperm motility / T=1 icosahedral viral capsid / receptor-mediated endocytosis of virus by host cell / trans-Golgi network / neuron migration / cytoplasmic vesicle / Golgi membrane / nucleolus / structural molecule activity / Golgi apparatus / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Adeno-associated virus - 5 Adeno-associated virus - 5 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Silveria M / Chapman MS | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2020 Journal: Viruses / Year: 2020Title: The Structure of an AAV5-AAVR Complex at 2.5 angstrom Resolution: Implications for Cellular Entry and Immune Neutralization of AAV Gene Therapy Vectors. Authors: Silveria MA / Large EE / Zane GM / White TA / Chapman MS | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22988.map.gz emd_22988.map.gz | 106.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22988-v30.xml emd-22988-v30.xml emd-22988.xml emd-22988.xml | 24.2 KB 24.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22988.png emd_22988.png | 260.9 KB | ||
| Masks |  emd_22988_msk_1.map emd_22988_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22988.cif.gz emd-22988.cif.gz | 7.3 KB | ||
| Others |  emd_22988_additional_1.map.gz emd_22988_additional_1.map.gz emd_22988_half_map_1.map.gz emd_22988_half_map_1.map.gz emd_22988_half_map_2.map.gz emd_22988_half_map_2.map.gz | 423.1 MB 432.7 MB 432.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22988 http://ftp.pdbj.org/pub/emdb/structures/EMD-22988 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22988 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22988 | HTTPS FTP |
-Validation report
| Summary document |  emd_22988_validation.pdf.gz emd_22988_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22988_full_validation.pdf.gz emd_22988_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_22988_validation.xml.gz emd_22988_validation.xml.gz | 19.5 KB | Display | |
| Data in CIF |  emd_22988_validation.cif.gz emd_22988_validation.cif.gz | 23 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22988 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22988 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22988 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22988 | HTTPS FTP |
-Related structure data
| Related structure data |  7kpnMC  7kp3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22988.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22988.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Adeno-associated virus serotype 5 bound to its cellular receptor AAVR, cryo-em sharpened map at 2.5 Angstroms resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
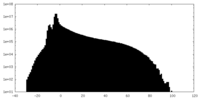
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66515 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
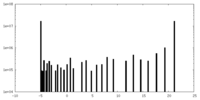
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22988_msk_1.map emd_22988_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
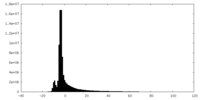
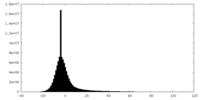
| Density Histograms |
-Additional map: Adeno-associated virus serotype 5 bound to its cellular...
| File | emd_22988_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Adeno-associated virus serotype 5 bound to its cellular receptor AAVR, cryo-em unsharpened map at 2.5 Angstroms resolution. | ||||||||||||
| Projections & Slices |
| ||||||||||||
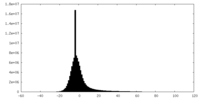
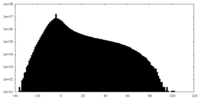
| Density Histograms |
-Half map: Adeno-associated virus serotype 5 bound to its cellular...
| File | emd_22988_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Adeno-associated virus serotype 5 bound to its cellular receptor AAVR, cryo-em half map 2 of 2 for a map at 2.5 Angstroms resolution. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Adeno-associated virus serotype 5 bound to its cellular...
| File | emd_22988_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Adeno-associated virus serotype 5 bound to its cellular receptor AAVR, cryo-em half map 1 of 2 for a map at 2.5 Angstroms resolution. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Adeno-associated virus - 5
| Entire | Name:  Adeno-associated virus - 5 Adeno-associated virus - 5 |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus - 5
| Supramolecule | Name: Adeno-associated virus - 5 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Expressed using SF9 cells with a pfastbac LIC vector. Purified with cesium chloride ultracentrifugation. NCBI-ID: 82300 / Sci species name: Adeno-associated virus - 5 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.746 MDa |
| Virus shell | Shell ID: 1 / Name: VP3 / Diameter: 250.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Dyslexia-associated protein KIAA0319-like protein
| Macromolecule | Name: Dyslexia-associated protein KIAA0319-like protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.182744 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASHHHHHHV SAGESVQITL PKNEVQLNAY VLQEPPKGET YTYDWQLITH PRDYSGEMEG KHSQILKLSK LTPGLYEFKV IVEGQNAHG EGYVNVTVKP EPRKNRPPIA IVSPQFQEIS LPTTSTVIDG SQSTDDDKIV QYHWEELKGP LREEKISEDT A ILKLSKLV ...String: MASHHHHHHV SAGESVQITL PKNEVQLNAY VLQEPPKGET YTYDWQLITH PRDYSGEMEG KHSQILKLSK LTPGLYEFKV IVEGQNAHG EGYVNVTVKP EPRKNRPPIA IVSPQFQEIS LPTTSTVIDG SQSTDDDKIV QYHWEELKGP LREEKISEDT A ILKLSKLV PGNYTFSLTV VDSDGATNST TANLTVNKAV D UniProtKB: Dyslexia-associated protein KIAA0319-like protein |
-Macromolecule #2: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Adeno-associated virus - 5 Adeno-associated virus - 5 |
| Molecular weight | Theoretical: 80.366211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SFVDHPPDWL EEVGEGLREF LGLEAGPPKP KPNQQHQDQA RGLVLPGYNY LGPGNGLDRG EPVNRADEVA REHDISYNEQ LEAGDNPYL KYNHADAEFQ EKLADDTSFG GNLGKAVFQA KKRVLEPFGL VEEGAKTAPT GKRIDDHFPK RKKARTEEDS K PSTSSDAE ...String: SFVDHPPDWL EEVGEGLREF LGLEAGPPKP KPNQQHQDQA RGLVLPGYNY LGPGNGLDRG EPVNRADEVA REHDISYNEQ LEAGDNPYL KYNHADAEFQ EKLADDTSFG GNLGKAVFQA KKRVLEPFGL VEEGAKTAPT GKRIDDHFPK RKKARTEEDS K PSTSSDAE AGPSGSQQLQ IPAQPASSLG ADTMSAGGGG PLGDNNQGAD GVGNASGDWH CDSTWMGDRV VTKSTRTWVL PS YNNHQYR EIKSGSVDGS NANAYFGYST PWGYFDFNRF HSHWSPRDWQ RLINNYWGFR PRSLRVKIFN IQVKEVTVQD STT TIANNL TSTVQVFTDD DYQLPYVVGN GTEGCLPAFP PQVFTLPQYG YATLNRDNTE NPTERSSFFC LEYFPSKMLR TGNN FEFTY NFEEVPFHSS FAPSQNLFKL ANPLVDQYLY RFVSTNNTGG VQFNKNLAGR YANTYKNWFP GPMGRTQGWN LGSGV NRAS VSAFATTNRM ELEGASYQVP PQPNGMTNNL QGSNTYALEN TMIFNSQPAN PGTTATYLEG NMLITSESET QPVNRV AYN VGGQMATNNQ SSTTAPATGT YNLQEIVPGS VWMERDVYLQ GPIWAKIPET GAHFHPSPAM GGFGLKHPPP MMLIKNT PV PGNITSFSDV PVSSFITQYS TGQVTVEMEW ELKKENSKRW NPEIQYTNNY NDPQFVDFAP DSTGEYRTTR PIGTRYLT R PL UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV Details: Two 2uL aliquots applied to grid (manual blotting between), prior to automated 3 second blot before plunging.. | ||||||||||||
| Details | Monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 93.0 K / Max: 93.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 1 / Number real images: 734 / Average electron dose: 32.9 e/Å2 / Details: Pixel size was 0.664 angstrom. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.7 µm / Nominal defocus min: -0.7000000000000001 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Stand-alone RSRef was used for refinement of magnification, resolution, envelope correction and atomic B-factors. This was alternated with RSRef-embedded CNS was used for molecular dynamics optimization (1st round) and stereochemically-restrained all-atom least-squares optimization. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Least-squares residual |
| Output model |  PDB-7kpn: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)