[English] 日本語
 Yorodumi
Yorodumi- EMDB-22473: Cryo-EM structure of unliganded octameric prenyltransferase domai... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22473 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of unliganded octameric prenyltransferase domain of Phomopsis amygdali fusicoccadiene synthase | |||||||||
 Map data Map data | Sharpened map of PaFS C-terminal prenyltransferase octamer with C2 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GGPP Synthase / Prenyltransferase / TRANSFERASE / LYASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationfusicocca-2,10(14)-diene synthase / alcohol biosynthetic process / mycotoxin biosynthetic process / geranylgeranyl diphosphate synthase / geranylgeranyl diphosphate synthase activity / isoprenoid biosynthetic process / lyase activity / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Phomopsis amygdali (fungus) Phomopsis amygdali (fungus) | |||||||||
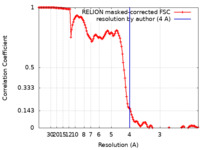
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Faylo JL / van Eeuwen T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural insight on assembly-line catalysis in terpene biosynthesis. Authors: Jacque L Faylo / Trevor van Eeuwen / Hee Jong Kim / Jose J Gorbea Colón / Benjamin A Garcia / Kenji Murakami / David W Christianson /  Abstract: Fusicoccadiene synthase from Phomopsis amygdali (PaFS) is a unique bifunctional terpenoid synthase that catalyzes the first two steps in the biosynthesis of the diterpene glycoside Fusicoccin A, a ...Fusicoccadiene synthase from Phomopsis amygdali (PaFS) is a unique bifunctional terpenoid synthase that catalyzes the first two steps in the biosynthesis of the diterpene glycoside Fusicoccin A, a mediator of 14-3-3 protein interactions. The prenyltransferase domain of PaFS generates geranylgeranyl diphosphate, which the cyclase domain then utilizes to generate fusicoccadiene, the tricyclic hydrocarbon skeleton of Fusicoccin A. Here, we use cryo-electron microscopy to show that the structure of full-length PaFS consists of a central octameric core of prenyltransferase domains, with the eight cyclase domains radiating outward via flexible linker segments in variable splayed-out positions. Cryo-electron microscopy and chemical crosslinking experiments additionally show that compact conformations can be achieved in which cyclase domains are more closely associated with the prenyltransferase core. This structural analysis provides a framework for understanding substrate channeling, since most of the geranylgeranyl diphosphate generated by the prenyltransferase domains remains on the enzyme for cyclization to form fusicoccadiene. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22473.map.gz emd_22473.map.gz | 192.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22473-v30.xml emd-22473-v30.xml emd-22473.xml emd-22473.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22473_fsc.xml emd_22473_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_22473.png emd_22473.png | 137.3 KB | ||
| Masks |  emd_22473_msk_1.map emd_22473_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22473.cif.gz emd-22473.cif.gz | 6.4 KB | ||
| Others |  emd_22473_additional_1.map.gz emd_22473_additional_1.map.gz emd_22473_half_map_1.map.gz emd_22473_half_map_1.map.gz emd_22473_half_map_2.map.gz emd_22473_half_map_2.map.gz | 108.5 MB 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22473 http://ftp.pdbj.org/pub/emdb/structures/EMD-22473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22473 | HTTPS FTP |
-Related structure data
| Related structure data |  7jthMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22473.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22473.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of PaFS C-terminal prenyltransferase octamer with C2 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.142 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
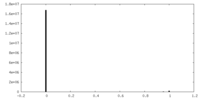
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22473_msk_1.map emd_22473_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
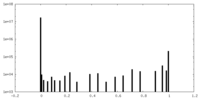
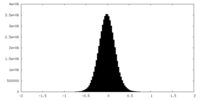
| Density Histograms |
-Additional map: Unsharpened, unfiltered reconstruction of PaFS C-terminal prenyltransferase octamer...
| File | emd_22473_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened, unfiltered reconstruction of PaFS C-terminal prenyltransferase octamer | ||||||||||||
| Projections & Slices |
| ||||||||||||
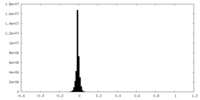
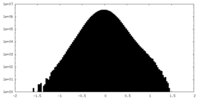
| Density Histograms |
-Half map: Unsharpened, unfiltered half-map of PaFS C-terminal prenyltransferase octamer...
| File | emd_22473_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened, unfiltered half-map of PaFS C-terminal prenyltransferase octamer with C2 symmetry | ||||||||||||
| Projections & Slices |
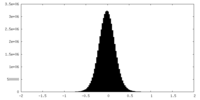
| ||||||||||||
| Density Histograms |
-Half map: Unsharpened, unfiltered half-map of PaFS C-terminal prenyltransferase octamer...
| File | emd_22473_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened, unfiltered half-map of PaFS C-terminal prenyltransferase octamer with C2 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
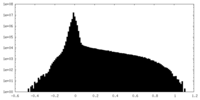
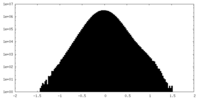
| Density Histograms |
- Sample components
Sample components
-Entire : Unliganded Phomopsis amygdali fusicoccadiene synthase octamer
| Entire | Name: Unliganded Phomopsis amygdali fusicoccadiene synthase octamer |
|---|---|
| Components |
|
-Supramolecule #1: Unliganded Phomopsis amygdali fusicoccadiene synthase octamer
| Supramolecule | Name: Unliganded Phomopsis amygdali fusicoccadiene synthase octamer type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Octamer of C-terminal prenyltransferase domain |
|---|---|
| Source (natural) | Organism:  Phomopsis amygdali (fungus) Phomopsis amygdali (fungus) |
| Molecular weight | Theoretical: 392 KDa |
-Macromolecule #1: Fusicoccadiene synthase
| Macromolecule | Name: Fusicoccadiene synthase / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: fusicocca-2,10(14)-diene synthase |
|---|---|
| Source (natural) | Organism:  Phomopsis amygdali (fungus) Phomopsis amygdali (fungus) |
| Molecular weight | Theoretical: 83.881891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MEFKYSEVVE PSTYYTEGLC EGIDVRKSKF TTLEDRGAIR AHEDWNKHIG PCGEYRGTLG PRFSFISVA VPECIPERLE VISYANEFAF LHDDVTDHVG HDTGEVENDE MMTVFLEAAH TGAIDTSNKV DIRRAGKKRI Q SQLFLEML ...String: MGSSHHHHHH SSGLVPRGSH MEFKYSEVVE PSTYYTEGLC EGIDVRKSKF TTLEDRGAIR AHEDWNKHIG PCGEYRGTLG PRFSFISVA VPECIPERLE VISYANEFAF LHDDVTDHVG HDTGEVENDE MMTVFLEAAH TGAIDTSNKV DIRRAGKKRI Q SQLFLEML AIDPECAKTT MKSWARFVEV GSSRQHETRF VELAKYIPYR IMDVGEMFWF GLVTFGLGLH IPDHELELCR EL MANAWIA VGLQNDIWSW PKERDAATLH GKDHVVNAIW VLMQEHQTDV DGAMQICRKL IVEYVAKYLE VIEATKNDES ISL DLRKYL DAMLYSISGN VVWSLECPRY NPDVSFNKTQ LEWMRQGLPS LESCPVLARS PEIDSDESAV SPTADESDST EDSL GSGSR QDSSLSTGLS LSPVHSNEGK DLQRVDTDHI FFEKAVLEAP YDYIASMPSK GVRDQFIDAL NDWLRVPDVK VGKIK DAVR VLHNSSLLLD DFQDNSPLRR GKPSTHNIFG SAQTVNTATY SIIKAIGQIM EFSAGESVQE VMNSIMILFQ GQAMDL FWT YNGHVPSEEE YYRMIDQKTG QLFSIATSLL LNAADNEIPR TKIQSCLHRL TRLLGRCFQI RDDYQNLVSA DYTKQKG FC EDLDEGKWSL ALIHMIHKQR SHMALLNVLS TGRKHGGMTL EQKQFVLDII EEEKSLDYTR SVMMDLHVQL RAEIGRIE I LLDSPNPAMR LLLELLRV UniProtKB: Fusicoccadiene synthase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 30.0 kPa / Details: 25 mA current was applied to grid. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 4.5 seconds before plunging.. | ||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 1 / Number real images: 1500 / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)