+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2163 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dodecameric human RuvBL1-RuvBL2 complex (compact conformation) | |||||||||
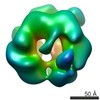
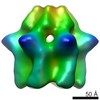
 Map data Map data | Reconstruction of the RuvBL1-RuvBL2 complex (compact conformation) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RuvBL1 / RuvBL2 / Rvb1 / Rvb2 / Pontin / Reptin / AAA+ | |||||||||
| Function / homology | PapC-like, C-terminal domain / NuA4 histone acetyltransferase complex Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 15.0 Å | |||||||||
 Authors Authors | Lopez-Perrote A / Munoz-Hernandez H / Gil D / Llorca O | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2012 Journal: Nucleic Acids Res / Year: 2012Title: Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex. Authors: Andrés López-Perrote / Hugo Muñoz-Hernández / David Gil / Oscar Llorca /  Abstract: RuvBL1 and RuvBL2, also known as Pontin and Reptin, are AAA+ proteins essential in small nucleolar ribonucloprotein biogenesis, chromatin remodelling, nonsense-mediated messenger RNA decay and ...RuvBL1 and RuvBL2, also known as Pontin and Reptin, are AAA+ proteins essential in small nucleolar ribonucloprotein biogenesis, chromatin remodelling, nonsense-mediated messenger RNA decay and telomerase assembly, among other functions. They are homologous to prokaryotic RuvB, forming single- and double-hexameric rings; however, a DNA binding domain II (DII) is inserted within the AAA+ core. Despite their biological significance, questions remain regarding their structure. Here, we report cryo-electron microscopy structures of human double-ring RuvBL1-RuvBL2 complexes at ∼15 Å resolution. Significantly, we resolve two coexisting conformations, compact and stretched, by image classification techniques. Movements in DII domains drive these conformational transitions, extending the complex and regulating the exposure of DNA binding regions. DII domains connect with the AAA+ core and bind nucleic acids, suggesting that these conformational changes could impact the regulation of RuvBL1-RuvBL2 containing complexes. These findings resolve some of the controversies in the structure of RuvBL1-RuvBL2 by revealing a mechanism that extends the complex by adjustments in DII. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2163.map.gz emd_2163.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2163-v30.xml emd-2163-v30.xml emd-2163.xml emd-2163.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2163.jpg emd_2163.jpg | 43.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2163 http://ftp.pdbj.org/pub/emdb/structures/EMD-2163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2163 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2163.map.gz / Format: CCP4 / Size: 4.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2163.map.gz / Format: CCP4 / Size: 4.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the RuvBL1-RuvBL2 complex (compact conformation) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
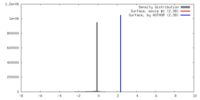
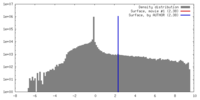
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Reconstruction of the human RuvBL1-RuvBL2 complex (compact confor...
| Entire | Name: Reconstruction of the human RuvBL1-RuvBL2 complex (compact conformation) |
|---|---|
| Components |
|
-Supramolecule #1000: Reconstruction of the human RuvBL1-RuvBL2 complex (compact confor...
| Supramolecule | Name: Reconstruction of the human RuvBL1-RuvBL2 complex (compact conformation) type: sample / ID: 1000 / Oligomeric state: Dodecameric / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: RuvBL1
| Macromolecule | Name: RuvBL1 / type: protein_or_peptide / ID: 1 / Name.synonym: Rvb1, Pontin, TIP49, TIP49a / Details: full length / Number of copies: 6 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: NuA4 histone acetyltransferase complex / InterPro: PapC-like, C-terminal domain |
-Macromolecule #2: RuvBL2
| Macromolecule | Name: RuvBL2 / type: protein_or_peptide / ID: 2 / Name.synonym: Rvb2, Reptin, TIP48, TIP49b / Details: full length / Number of copies: 6 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: NuA4 histone acetyltransferase complex |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 250mM NaCl, 25 mM Tris-HCL |
| Grid | Details: Quantifoil grids 300 mesh R2/1 holey carbon copper grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III Method: Manual application (4 microliters) Blot offset: -2 mm Blot total:2 Blot time: 2s Wait time: 30s Drain time: 1s |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Temperature | Min: 83 K / Max: 100 K / Average: 91.5 K |
| Alignment procedure | Legacy - Astigmatism: Phase flipping (Objective lens astigmatism was corrected using the CCD and the power spectrum) |
| Specialist optics | Energy filter - Name: In-column nergy filter (Omega Filter) / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 10.0 eV |
| Date | Mar 28, 2012 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 338 / Average electron dose: 12 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 86855 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each CCD Frame, estimated with CTFFIND and corrected using BSOFT |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D6 (2x6 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 15.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN1.9, EMAN2, XMIPP, BSOFT / Number images used: 12740 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid Body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross correlation |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid Body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross correlation |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)