[English] 日本語
 Yorodumi
Yorodumi- EMDB-21356: Structure of a bacterial Atm1-family ABC exporter with MgADPVO4 bound -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21356 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a bacterial Atm1-family ABC exporter with MgADPVO4 bound | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / membrane protein / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTranslocases / response to mercury ion / ABC-type transporter activity / monoatomic ion transport / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Novosphingobium aromaticivorans DSM 12444 (bacteria) / Novosphingobium aromaticivorans DSM 12444 (bacteria) /  Novosphingobium aromaticivorans (strain ATCC 700278 / DSM 12444 / CIP 105152 / NBRC 16084 / F199) (bacteria) Novosphingobium aromaticivorans (strain ATCC 700278 / DSM 12444 / CIP 105152 / NBRC 16084 / F199) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.03 Å | |||||||||
 Authors Authors | Fan C / Rees DC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: A structural framework for unidirectional transport by a bacterial ABC exporter. Authors: Chengcheng Fan / Jens T Kaiser / Douglas C Rees /  Abstract: The ATP-binding cassette (ABC) transporter of mitochondria (Atm1) mediates iron homeostasis in eukaryotes, while the prokaryotic homolog from (Atm1) can export glutathione derivatives and confer ...The ATP-binding cassette (ABC) transporter of mitochondria (Atm1) mediates iron homeostasis in eukaryotes, while the prokaryotic homolog from (Atm1) can export glutathione derivatives and confer protection against heavy-metal toxicity. To establish the structural framework underlying the Atm1 transport mechanism, we determined eight structures by X-ray crystallography and single-particle cryo-electron microscopy in distinct conformational states, stabilized by individual disulfide crosslinks and nucleotides. As Atm1 progresses through the transport cycle, conformational changes in transmembrane helix 6 (TM6) alter the glutathione-binding site and the associated substrate-binding cavity. Significantly, kinking of TM6 in the post-ATP hydrolysis state stabilized by MgADPVO eliminates this cavity, precluding uptake of glutathione derivatives. The presence of this cavity during the transition from the inward-facing to outward-facing conformational states, and its absence in the reverse direction, thereby provide an elegant and conceptually simple mechanism for enforcing the export directionality of transport by Atm1. One of the disulfide crosslinked Atm1 variants characterized in this work retains significant glutathione transport activity, suggesting that ATP hydrolysis and substrate transport by Atm1 may involve a limited set of conformational states with minimal separation of the nucleotide-binding domains in the inward-facing conformation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21356.map.gz emd_21356.map.gz | 51.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21356-v30.xml emd-21356-v30.xml emd-21356.xml emd-21356.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21356.png emd_21356.png | 134.7 KB | ||
| Filedesc metadata |  emd-21356.cif.gz emd-21356.cif.gz | 6.2 KB | ||
| Others |  emd_21356_additional.map.gz emd_21356_additional.map.gz emd_21356_half_map_1.map.gz emd_21356_half_map_1.map.gz emd_21356_half_map_2.map.gz emd_21356_half_map_2.map.gz | 97.1 MB 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21356 http://ftp.pdbj.org/pub/emdb/structures/EMD-21356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21356 | HTTPS FTP |
-Validation report
| Summary document |  emd_21356_validation.pdf.gz emd_21356_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21356_full_validation.pdf.gz emd_21356_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_21356_validation.xml.gz emd_21356_validation.xml.gz | 13.4 KB | Display | |
| Data in CIF |  emd_21356_validation.cif.gz emd_21356_validation.cif.gz | 15.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21356 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21356 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21356 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21356 | HTTPS FTP |
-Related structure data
| Related structure data |  6vqtMC  6pamC  6panC  6paoC  6paqC  6parC  6vquC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21356.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21356.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8522 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
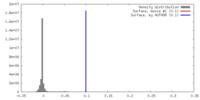
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened map
| File | emd_21356_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
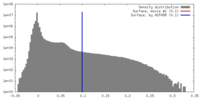
| Density Histograms |
-Half map: #2
| File | emd_21356_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
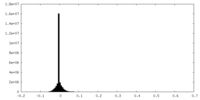
| Density Histograms |
-Half map: #1
| File | emd_21356_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
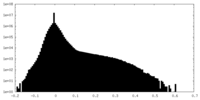
| Density Histograms |
- Sample components
Sample components
-Entire : homodimeric ATM1-type heavy metal exporter
| Entire | Name: homodimeric ATM1-type heavy metal exporter |
|---|---|
| Components |
|
-Supramolecule #1: homodimeric ATM1-type heavy metal exporter
| Supramolecule | Name: homodimeric ATM1-type heavy metal exporter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Novosphingobium aromaticivorans DSM 12444 (bacteria) Novosphingobium aromaticivorans DSM 12444 (bacteria) |
| Molecular weight | Theoretical: 133 KDa |
-Macromolecule #1: ATM1-type heavy metal exporter
| Macromolecule | Name: ATM1-type heavy metal exporter / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Novosphingobium aromaticivorans (strain ATCC 700278 / DSM 12444 / CIP 105152 / NBRC 16084 / F199) (bacteria) Novosphingobium aromaticivorans (strain ATCC 700278 / DSM 12444 / CIP 105152 / NBRC 16084 / F199) (bacteria)Strain: ATCC 700278 / DSM 12444 / CIP 105152 / NBRC 16084 / F199 |
| Molecular weight | Theoretical: 67.771602 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPPETATNPK DARHDGWQTL KRFLPYLWPA DNAVLRRRVV GAILMVLLGK ATTLALPFAY KKAVDAMTLG GGAQPALTVA LAFVLAYAL GRFSGVLFDN LRNIVFERVG QDATRHLAEN VFARLHKLSL RFHLARRTGE VTKVIERGTK SIDTMLYFLL F NIAPTVIE ...String: MPPETATNPK DARHDGWQTL KRFLPYLWPA DNAVLRRRVV GAILMVLLGK ATTLALPFAY KKAVDAMTLG GGAQPALTVA LAFVLAYAL GRFSGVLFDN LRNIVFERVG QDATRHLAEN VFARLHKLSL RFHLARRTGE VTKVIERGTK SIDTMLYFLL F NIAPTVIE LTAVIVIFWL NFGLGLVTAT ILAVIAYVWT TRTITEWRTH LREKMNRLDG QALARAVDSL LNYETVKYFG AE SREEARY ASAARAYADA AVKSENSLGL LNIAQALIVN LLMAGAMAWT VYGWSQGKLT VGDLVFVNTY LTQLFRPLDM LGM VYRTIR QGLIDMAEMF RLIDTHIEVA DVPNAPALVV NRPSVTFDNV VFGYDRDREI LHGLSFEVAA GSRVAIVGPS GAGK STIAR LLFRFYDPWE GRILIDGQDI AHVTQTSLRA ALGIVPQDSV LFNDTIGYNI AYGRDGASRA EVDAAAKGAA IADFI ARLP QGYDTEVGER GLKLSGGEKQ RVAIARTLVK NPPILLFDEA TSALDTRTEQ DILSTMRAVA SHRTTISIAH RLSTIA DSD TILVLDQGRL AEQGSHLDLL RRDGLYAEMW ARQAAESAEV SEAAEHHHHH H UniProtKB: ATM1-type heavy metal exporter |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: VANADATE ION
| Macromolecule | Name: VANADATE ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: VO4 |
|---|---|
| Molecular weight | Theoretical: 114.939 Da |
| Chemical component information |  ChemComp-VN3: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | The sample was reconstituted with MSP1D1 nanodiscs and POPC. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)