[English] 日本語
 Yorodumi
Yorodumi- EMDB-21152: Tetradecameric PilQ bound by TsaP heptamer from Pseudomonas aeruginosa -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21152 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tetradecameric PilQ bound by TsaP heptamer from Pseudomonas aeruginosa | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type IV pilus / T4P / PilQ / TsaP / secretin / pilotin / type IVa pilus / T4aP / pilus / outer membrane / periplasm / bacterial secretion system / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | McCallum M / Tammam S | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP. Authors: Matthew McCallum / Stephanie Tammam / John L Rubinstein / Lori L Burrows / P Lynne Howell /  Abstract: The type IV pilus machinery is a multi-protein complex that polymerizes and depolymerizes a pilus fiber used for attachment, twitching motility, phage adsorption, natural competence, protein ...The type IV pilus machinery is a multi-protein complex that polymerizes and depolymerizes a pilus fiber used for attachment, twitching motility, phage adsorption, natural competence, protein secretion, and surface-sensing. An outer membrane secretin pore is required for passage of the pilus fiber out of the cell. Herein, the structure of the tetradecameric secretin, PilQ, from the Pseudomonas aeruginosa type IVa pilus system was determined to 4.3 Å and 4.4 Å resolution in the presence and absence of C symmetric spikes, respectively. The heptameric spikes were found to be two tandem C-terminal domains of TsaP. TsaP forms a belt around PilQ and while it is not essential for twitching motility, overexpression of TsaP triggers a signal cascade upstream of PilY1 leading to cyclic di-GMP up-regulation. These results resolve the identity of the spikes identified with Proteobacterial PilQ homologs and may reveal a new component of the surface-sensing cyclic di-GMP signal cascade. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21152.map.gz emd_21152.map.gz | 59.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21152-v30.xml emd-21152-v30.xml emd-21152.xml emd-21152.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21152.png emd_21152.png | 186.4 KB | ||
| Filedesc metadata |  emd-21152.cif.gz emd-21152.cif.gz | 6.1 KB | ||
| Others |  emd_21152_additional_1.map.gz emd_21152_additional_1.map.gz emd_21152_half_map_1.map.gz emd_21152_half_map_1.map.gz emd_21152_half_map_2.map.gz emd_21152_half_map_2.map.gz | 31.6 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21152 http://ftp.pdbj.org/pub/emdb/structures/EMD-21152 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21152 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21152 | HTTPS FTP |
-Related structure data
| Related structure data |  6ve2MC  6ve3C  6ve4C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21152.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21152.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
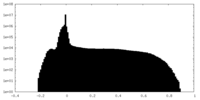
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened map
| File | emd_21152_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
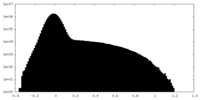
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A
| File | emd_21152_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_21152_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : PilQ with TsaP
| Entire | Name: PilQ with TsaP |
|---|---|
| Components |
|
-Supramolecule #1: PilQ with TsaP
| Supramolecule | Name: PilQ with TsaP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Affinity purified PilQ with pulled-down TsaP |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
-Macromolecule #1: Fimbrial assembly protein PilQ
| Macromolecule | Name: Fimbrial assembly protein PilQ / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 79.731141 KDa |
| Recombinant expression | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Sequence | String: MNSGLSRLGI ALLAAMFAPA LLAADLEKLD VAALPGDRVE LKLQFDEPVA APRGYTIEQP ARIALDLPGV QNKLGTKNRE LSVGNTRSV TVVEAKDRTR LIINLTALSS YTTRVEGNNL FVVVGNSPHH HHHHHHAGAS VASAAPVKAS PAPASYAQPI K PKPYVPAG ...String: MNSGLSRLGI ALLAAMFAPA LLAADLEKLD VAALPGDRVE LKLQFDEPVA APRGYTIEQP ARIALDLPGV QNKLGTKNRE LSVGNTRSV TVVEAKDRTR LIINLTALSS YTTRVEGNNL FVVVGNSPHH HHHHHHAGAS VASAAPVKAS PAPASYAQPI K PKPYVPAG RAIRNIDFQR GEKGEGNVVI DLSDPTLSPD IQEQGGKIRL DFAKTQLPDA LRVRLDVKDF ATPVQFVNAS AQ SDRTSIT IEPSGLYDYL VYQTDNRLTV SIKPMTTEDA ERRKKDNFAY TGEKLSLNFQ DIDVRSVLQL IADFTDLNLV ASD TVQGNI TLRLQNVPWD QALDLVLKTK GLDKRKLGNV LLVAPADEIA ARERQELEAQ KQIAELAPLR RELIQVNYAK AADI AKLFQ SVTSDGGQEG KEGGRGSITV DDRTNSIIAY QPQERLDELR RIVSQLDIPV RQVMIEARIV EANVGYDKSL GVRWG GAYH KGNWSGYGKD GNIGIKDEDG MNCGPIAGSC TFPTTGTSKS PSPFVDLGAK DATSGIGIGF ITDNIILDLQ LSAMEK TGN GEIVSQPKVV TSDKETAKIL KGSEVPYQEA SSSGATSTSF KEAALSLEVT PQITPDNRII VEVKVTKDAP DYQNMLN GV PPINKNEVNA KILVNDGETI VIGGVFSNEQ SKSVEKVPFL GELPYLGRLF RRDTVTDRKN ELLVFLTPRI MNNQAIAI G RGHHHHHHHH UniProtKB: Fimbrial assembly protein PilQ |
-Macromolecule #2: LysM domain-containing protein
| Macromolecule | Name: LysM domain-containing protein / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 37.632902 KDa |
| Recombinant expression | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Sequence | String: MRKSLVALLL LAASGLAQAQ VDLREGHPDR YTVVRGDTLW DISGKFLRQP WKWPELWHAN PQIQNPHLIY PGDTLSLVYV DGQPRLVLN RGESRGTIKL SPKIRSTPIA EAIPTIPLDK INSFLLANRI VDDEKTFTSA PYIVAGNAER IVSGTGDRIY A RGKFADGQ ...String: MRKSLVALLL LAASGLAQAQ VDLREGHPDR YTVVRGDTLW DISGKFLRQP WKWPELWHAN PQIQNPHLIY PGDTLSLVYV DGQPRLVLN RGESRGTIKL SPKIRSTPIA EAIPTIPLDK INSFLLANRI VDDEKTFTSA PYIVAGNAER IVSGTGDRIY A RGKFADGQ PAYGIFRQGK VYIDPKTKEV LGINADDIGG GEVVATEGDV ATLALTRTTQ EVRLGDRLFP TEERAVNSTF MP GEPSREV KGEIIDVPRG VTQIGQFDVV TLNRGQRDGL AEGNVLAIYK VGETVRDRVT GESVKIPDER AGLLMVFRTY KKL SYALVL MASRPLSVTD RVQNP UniProtKB: LysM domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 35.7 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL In silico model: Starting models generated in Phyre2 and I-TASSER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C7 (7 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 4.3 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 47468 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 98 |
|---|---|
| Output model |  PDB-6ve2: |
 Movie
Movie Controller
Controller








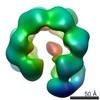


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)