[English] 日本語
 Yorodumi
Yorodumi- EMDB-21043: Structure of human 2E01 Fab in complex with influenza virus neura... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21043 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human 2E01 Fab in complex with influenza virus neuraminidase from B/Phuket/3073/2013 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | viral protein / neuraminidase / Structural Genomics / Center for Structural Genomics of Infectious Diseases / CSGID | |||||||||
| Function / homology |  Function and homology information Function and homology informationexo-alpha-sialidase / exo-alpha-sialidase activity / carbohydrate metabolic process / host cell plasma membrane / virion membrane / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Influenza B virus / Influenza B virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
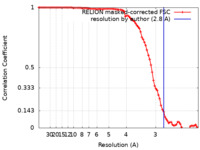
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Dai YN / Fremont DH | |||||||||
 Citation Citation |  Journal: Immunity / Year: 2020 Journal: Immunity / Year: 2020Title: Human Antibodies Targeting Influenza B Virus Neuraminidase Active Site Are Broadly Protective. Authors: Anders Madsen / Ya-Nan Dai / Meagan McMahon / Aaron J Schmitz / Jackson S Turner / Jessica Tan / Tingting Lei / Wafaa B Alsoussi / Shirin Strohmeier / Mostafa Amor / Bassem M Mohammed / ...Authors: Anders Madsen / Ya-Nan Dai / Meagan McMahon / Aaron J Schmitz / Jackson S Turner / Jessica Tan / Tingting Lei / Wafaa B Alsoussi / Shirin Strohmeier / Mostafa Amor / Bassem M Mohammed / Philip A Mudd / Viviana Simon / Rebecca J Cox / Daved H Fremont / Florian Krammer / Ali H Ellebedy /   Abstract: Influenza B virus (IBV) infections can cause severe disease in children and the elderly. Commonly used antivirals have lower clinical effectiveness against IBV compared to influenza A viruses (IAV). ...Influenza B virus (IBV) infections can cause severe disease in children and the elderly. Commonly used antivirals have lower clinical effectiveness against IBV compared to influenza A viruses (IAV). Neuraminidase (NA), the second major surface protein on the influenza virus, is emerging as a target of broadly protective antibodies that recognize the NA active site of IAVs. However, similarly broadly protective antibodies against IBV NA have not been identified. Here, we isolated and characterized human monoclonal antibodies (mAbs) that target IBV NA from an IBV-infected patient. Two mAbs displayed broad and potent capacity to inhibit IBV NA enzymatic activity, neutralize the virus in vitro, and protect against lethal IBV infection in mice in prophylactic and therapeutic settings. These mAbs inserted long CDR-H3 loops into the NA active site, engaging residues highly conserved among IBV NAs. These mAbs provide a blueprint for the development of improved vaccines and therapeutics against IBVs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21043.map.gz emd_21043.map.gz | 55.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21043-v30.xml emd-21043-v30.xml emd-21043.xml emd-21043.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21043_fsc.xml emd_21043_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_21043.png emd_21043.png | 26.6 KB | ||
| Filedesc metadata |  emd-21043.cif.gz emd-21043.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21043 http://ftp.pdbj.org/pub/emdb/structures/EMD-21043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21043 | HTTPS FTP |
-Related structure data
| Related structure data |  6v4oMC  6v4nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21043.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21043.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Structure of human 2E01 Fab in complex with influenza virus neura...
| Entire | Name: Structure of human 2E01 Fab in complex with influenza virus neuraminidase from B/Phuket/3073/2013 |
|---|---|
| Components |
|
-Supramolecule #1: Structure of human 2E01 Fab in complex with influenza virus neura...
| Supramolecule | Name: Structure of human 2E01 Fab in complex with influenza virus neuraminidase from B/Phuket/3073/2013 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 400 KDa |
-Supramolecule #2: Influenza virus neuraminidase
| Supramolecule | Name: Influenza virus neuraminidase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Influenza B virus / Strain: B/Phuket/3073/2013 Influenza B virus / Strain: B/Phuket/3073/2013 |
-Supramolecule #3: Human antibody
| Supramolecule | Name: Human antibody / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Neuraminidase
| Macromolecule | Name: Neuraminidase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: exo-alpha-sialidase |
|---|---|
| Source (natural) | Organism:  Influenza B virus Influenza B virus |
| Molecular weight | Theoretical: 50.038824 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: ADPHHHHHHS SSDYSDLQRV KQELLEEVKK ELQKVKEEII EAFVQELRKR GSLVPRGSPS RWTYPRLSCP GSTFQKALLI SPHRFGETK GNSAPLIIRE PFIACGPKEC KHFALTHYAA QPGGYYNGTR EDRNKLRHLI SVKLGKIPTV ENSIFHMAAW S GSACHDGR ...String: ADPHHHHHHS SSDYSDLQRV KQELLEEVKK ELQKVKEEII EAFVQELRKR GSLVPRGSPS RWTYPRLSCP GSTFQKALLI SPHRFGETK GNSAPLIIRE PFIACGPKEC KHFALTHYAA QPGGYYNGTR EDRNKLRHLI SVKLGKIPTV ENSIFHMAAW S GSACHDGR EWTYIGVDGP DSNALLKIKY GEAYTDTYHS YAKNILRTQE SACNCIGGDC YLMITDGPAS GISECRFLKI RE GRIIKEI FPTGRVKHTE ECTCGFASNK TIECACRDNS YTAKRPFVKL NVETDTAEIR LMCTKTYLDT PRPNDGSITG PCE SDGDEG SGGIKGGFVH QRMASKIGRW YSRTMSKTKR MGMGLYVKYD GDPWTDSEAL ALSGVMVSME EPGWYSFGFE IKDK KCDVP CIGIEMVHDG GKTTWHSAAT AIYCLMGSGQ LLWDTVTGVN MTL UniProtKB: Neuraminidase |
-Macromolecule #2: Antibody Fab heavy chain
| Macromolecule | Name: Antibody Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.840137 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVQSGAE VKKPGSSVKV SCKASGYTFI NHALSWVRQA PGQGLEWVGG IIPIFGLAKY GQKFQDRVTI TADESTKTAY MDLRSLRSD DTAVYYCARD TVAVYEDFDW SSPYFFYMDV WGKGTTVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT ...String: EVQLVQSGAE VKKPGSSVKV SCKASGYTFI NHALSWVRQA PGQGLEWVGG IIPIFGLAKY GQKFQDRVTI TADESTKTAY MDLRSLRSD DTAVYYCARD TVAVYEDFDW SSPYFFYMDV WGKGTTVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCRSLVP RG SSGHHHH HH |
-Macromolecule #3: Antibody Fab light chain
| Macromolecule | Name: Antibody Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.327992 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPAT LSLFPGERAT LSCRASQSAG SKSLAWYQHK VGQPPRLLIN GASSRATGIP DRFSGSGSGP DFNLTISRLE PEDFAVYYC QRYGTSLVTF GGGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: EIVLTQSPAT LSLFPGERAT LSCRASQSAG SKSLAWYQHK VGQPPRLLIN GASSRATGIP DRFSGSGSGP DFNLTISRLE PEDFAVYYC QRYGTSLVTF GGGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #5: beta-D-mannopyranose
| Macromolecule | Name: beta-D-mannopyranose / type: ligand / ID: 5 / Number of copies: 4 / Formula: BMA |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-BMA: |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 89 / Target criteria: CORRELATION COEFFICIENT |
| Output model |  PDB-6v4o: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)