[English] 日本語
 Yorodumi
Yorodumi- EMDB-20822: Human metabotropic GABA(B) receptor bound to agonist SKF97541 and... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20822 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
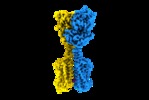
| Title | Human metabotropic GABA(B) receptor bound to agonist SKF97541 and positive allosteric modulator GS39783. | |||||||||
 Map data Map data | Sharpened map of GABAB heterodimer in complex with agonist (SKF97541) and PAM (GS39783). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G protein-coupled receptor / GABA / GABAB / PAM / Neurotransmitter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential / GABA B receptor activation / G protein-coupled neurotransmitter receptor activity involved in regulation of presynaptic membrane potential / G protein-coupled GABA receptor complex / negative regulation of gamma-aminobutyric acid secretion / neuron-glial cell signaling / G protein-coupled GABA receptor activity / G protein-coupled receptor heterodimeric complex / negative regulation of epinephrine secretion / negative regulation of dopamine secretion ...G protein-coupled neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential / GABA B receptor activation / G protein-coupled neurotransmitter receptor activity involved in regulation of presynaptic membrane potential / G protein-coupled GABA receptor complex / negative regulation of gamma-aminobutyric acid secretion / neuron-glial cell signaling / G protein-coupled GABA receptor activity / G protein-coupled receptor heterodimeric complex / negative regulation of epinephrine secretion / negative regulation of dopamine secretion / positive regulation of growth hormone secretion / extracellular matrix protein binding / GABA receptor complex / negative regulation of adenylate cyclase activity / positive regulation of glutamate secretion / Class C/3 (Metabotropic glutamate/pheromone receptors) / synaptic transmission, GABAergic / gamma-aminobutyric acid signaling pathway / negative regulation of synaptic transmission / axolemma / GABA-ergic synapse / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / dendritic shaft / response to nicotine / mitochondrial membrane / Schaffer collateral - CA1 synapse / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / osteoblast differentiation / transmembrane signaling receptor activity / synaptic vesicle / presynaptic membrane / G alpha (i) signalling events / chemical synaptic transmission / postsynaptic membrane / response to ethanol / dendritic spine / neuron projection / protein heterodimerization activity / G protein-coupled receptor signaling pathway / negative regulation of cell population proliferation / neuronal cell body / glutamatergic synapse / endoplasmic reticulum membrane / extracellular space / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.63 Å | |||||||||
 Authors Authors | Shaye H / Han GW | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structural basis of the activation of a metabotropic GABA receptor. Authors: Hamidreza Shaye / Andrii Ishchenko / Jordy Homing Lam / Gye Won Han / Li Xue / Philippe Rondard / Jean-Philippe Pin / Vsevolod Katritch / Cornelius Gati / Vadim Cherezov /   Abstract: Metabotropic γ-aminobutyric acid receptors (GABA) are involved in the modulation of synaptic responses in the central nervous system and have been implicated in neuropsychological conditions that ...Metabotropic γ-aminobutyric acid receptors (GABA) are involved in the modulation of synaptic responses in the central nervous system and have been implicated in neuropsychological conditions that range from addiction to psychosis. GABA belongs to class C of the G-protein-coupled receptors, and its functional entity comprises an obligate heterodimer that is composed of the GB1 and GB2 subunits. Each subunit possesses an extracellular Venus flytrap domain, which is connected to a canonical seven-transmembrane domain. Here we present four cryo-electron microscopy structures of the human full-length GB1-GB2 heterodimer: one structure of its inactive apo state, two intermediate agonist-bound forms and an active form in which the heterodimer is bound to an agonist and a positive allosteric modulator. The structures reveal substantial differences, which shed light on the complex motions that underlie the unique activation mechanism of GABA. Our results show that agonist binding leads to the closure of the Venus flytrap domain of GB1, triggering a series of transitions, first rearranging and bringing the two transmembrane domains into close contact along transmembrane helix 6 and ultimately inducing conformational rearrangements in the GB2 transmembrane domain via a lever-like mechanism to initiate downstream signalling. This active state is stabilized by a positive allosteric modulator binding at the transmembrane dimerization interface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20822.map.gz emd_20822.map.gz | 129.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20822-v30.xml emd-20822-v30.xml emd-20822.xml emd-20822.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20822.png emd_20822.png | 96.3 KB | ||
| Filedesc metadata |  emd-20822.cif.gz emd-20822.cif.gz | 7.2 KB | ||
| Others |  emd_20822_additional.map.gz emd_20822_additional.map.gz | 68 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20822 http://ftp.pdbj.org/pub/emdb/structures/EMD-20822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20822 | HTTPS FTP |
-Validation report
| Summary document |  emd_20822_validation.pdf.gz emd_20822_validation.pdf.gz | 526.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20822_full_validation.pdf.gz emd_20822_full_validation.pdf.gz | 526.3 KB | Display | |
| Data in XML |  emd_20822_validation.xml.gz emd_20822_validation.xml.gz | 6.7 KB | Display | |
| Data in CIF |  emd_20822_validation.cif.gz emd_20822_validation.cif.gz | 7.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20822 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20822 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20822 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20822 | HTTPS FTP |
-Related structure data
| Related structure data |  6uo8MC  6uo9C  6uoaC  6vjmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20822.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20822.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of GABAB heterodimer in complex with agonist (SKF97541) and PAM (GS39783). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8521 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unfiltered map of GABAB heterodimer in complex with...
| File | emd_20822_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered map of GABAB heterodimer in complex with agonist (SKF97541) and PAM (GS39783). | ||||||||||||
| Projections & Slices |
| ||||||||||||
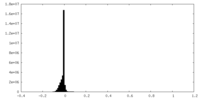
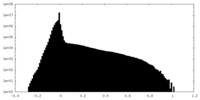
| Density Histograms |
- Sample components
Sample components
-Entire : GABA(B) receptor bound to agonist and PAM
| Entire | Name: GABA(B) receptor bound to agonist and PAM |
|---|---|
| Components |
|
-Supramolecule #1: GABA(B) receptor bound to agonist and PAM
| Supramolecule | Name: GABA(B) receptor bound to agonist and PAM / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Location in cell: Membrane Homo sapiens (human) / Location in cell: Membrane |
| Molecular weight | Theoretical: 174 KDa |
-Macromolecule #1: Gamma-aminobutyric acid type B receptor subunit 1
| Macromolecule | Name: Gamma-aminobutyric acid type B receptor subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 86.450977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSERRAVYIG ALFPMSGGWP GGQACQPAVE MALEDVNSRR DILPDYELKL IHHDSKCDPG QATKYLYELL YNDPIKIILM PGCSSVSTL VAEAARMWNL IVLSYGSSSP ALSNRQRFPT FFRTHPSATL HNPTRVKLFE KWGWKKIATI QQTTEVFTST L DDLEERVK ...String: GSERRAVYIG ALFPMSGGWP GGQACQPAVE MALEDVNSRR DILPDYELKL IHHDSKCDPG QATKYLYELL YNDPIKIILM PGCSSVSTL VAEAARMWNL IVLSYGSSSP ALSNRQRFPT FFRTHPSATL HNPTRVKLFE KWGWKKIATI QQTTEVFTST L DDLEERVK EAGIEITFRQ SFFSDPAVPV KNLKRQDARI IVGLFYETEA RKVFCEVYKE RLFGKKYVWF LIGWYADNWF KI YDPSINC TVDEMTEAVE GHITTEIVML NPANTRSISN MTSQEFVEKL TKRLKRHPEE TGGFQEAPLA YDAIWALALA LNK TSGGGG RSGVRLEDFN YNNQTITDQI YRAMNSSSFE GVSGHVVFDA SGSRMAWTLI EQLQGGSYKK IGYYDSTKDD LSWS KTDKW IGGSPPADQT LVIKTFRFLS QKLFISVSVL SSLGIVLAVV CLSFNIYNSH VRYIQNSQPN LNNLTAVGCS LALAA VFPL GLDGYHIGRN QFPFVCQARL WLLGLGFSLG YGSMFTKIWW VHTVFTKKEE KKEWRKTLEP WKLYATVGLL VGMDVL TLA IWQIVDPLHR TIETFAKEEP KEDIDVSILP QLEHCSSRKM NTWLGIFYGY KGLLLLLGIF LAYETKSVST EKINDHR AV GMAIYNVAVL CLITAPVTMI LSSQQDAAFA FASLAIVFSS YITLVVLFVP KMRRLITRGE WQSEAQDTMK TGSSTNNN E EEKSRLLEKE NRELEKIIAE KEERVSELRH QLQSRLEVLF Q UniProtKB: Gamma-aminobutyric acid type B receptor subunit 1 |
-Macromolecule #2: Gamma-aminobutyric acid type B receptor subunit 2
| Macromolecule | Name: Gamma-aminobutyric acid type B receptor subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 88.200844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GWARGAPRPP PSSPPLSIMG LMPLTKEVAK GSIGRGVLPA VELAIEQIRN ESLLRPYFLD LRLYDTECDN AKGLKAFYDA IKYGPNHLM VFGGVCPSVT SIIAESLQGW NLVQLSFAAT TPVLADKKKY PYFFRTVPSD NAVNPAILKL LKHYQWKRVG T LTQDVQRF ...String: GWARGAPRPP PSSPPLSIMG LMPLTKEVAK GSIGRGVLPA VELAIEQIRN ESLLRPYFLD LRLYDTECDN AKGLKAFYDA IKYGPNHLM VFGGVCPSVT SIIAESLQGW NLVQLSFAAT TPVLADKKKY PYFFRTVPSD NAVNPAILKL LKHYQWKRVG T LTQDVQRF SEVRNDLTGV LYGEDIEISD TESFSNDPCT SVKKLKGNDV RIILGQFDQN MAAKVFCCAY EENMYGSKYQ WI IPGWYEP SWWEQVHTEA NSSRCLRKNL LAAMEGYIGV DFEPLSSKQI KTISGKTPQQ YEREYNNKRS GVGPSKFHGY AYD GIWVIA KTLQRAMETL HASSRHQRIQ DFNYTDHTLG RIILNAMNET NFFGVTGQVV FRNGERMGTI KFTQFQDSRE VKVG EYNAV ADTLEIINDT IRFQGSEPPK DKTIILEQLR KISLPLYSIL SALTILGMIM ASAFLFFNIK NRNQKLIKMS SPYMN NLII LGGMLSYASI FLFGLDGSFV SEKTFETLCT VRTWILTVGY TTAFGAMFAK TWRVHAIFKN VKMKKKIIKD QKLLVI VGG MLLIDLCILI CWQAVDPLRR TVEKYSMEPD PAGRDISIRP LLEHCENTHM TIWLGIVYAY KGLLMLFGCF LAWETRN VS IPALNDSKYI GMSVYNVGIM CIIGAAVSFL TRDQPNVQFC IVALVIIFCS TITLCLVFVP KLITLRTNPD AATQNRRF Q FTQNQKKEDS KTSTSVTSVN QASTSRLEGL QSENHRLRMK ITELDKDLEE VTMQLQDT UniProtKB: Gamma-aminobutyric acid type B receptor subunit 2 |
-Macromolecule #5: (R)-(3-aminopropyl)methylphosphinic acid
| Macromolecule | Name: (R)-(3-aminopropyl)methylphosphinic acid / type: ligand / ID: 5 / Number of copies: 1 / Formula: QD7 |
|---|---|
| Molecular weight | Theoretical: 137.117 Da |
| Chemical component information | 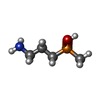 ChemComp-QD7: |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: N~4~,N~6~-dicyclopentyl-2-(methylsulfanyl)-5-nitropyrimidine-4,6-...
| Macromolecule | Name: N~4~,N~6~-dicyclopentyl-2-(methylsulfanyl)-5-nitropyrimidine-4,6-diamine type: ligand / ID: 7 / Number of copies: 1 / Formula: QDA |
|---|---|
| Molecular weight | Theoretical: 337.44 Da |
| Chemical component information | 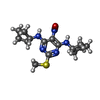 ChemComp-QDA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 10917 / Average exposure time: 0.07 sec. / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 29000 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.5 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-6uo8: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)