+Search query
-Structure paper
| Title | Structural basis of the activation of a metabotropic GABA receptor. |
|---|---|
| Journal, issue, pages | Nature, Vol. 584, Issue 7820, Page 298-303, Year 2020 |
| Publish date | Jun 17, 2020 |
 Authors Authors | Hamidreza Shaye / Andrii Ishchenko / Jordy Homing Lam / Gye Won Han / Li Xue / Philippe Rondard / Jean-Philippe Pin / Vsevolod Katritch / Cornelius Gati / Vadim Cherezov /   |
| PubMed Abstract | Metabotropic γ-aminobutyric acid receptors (GABA) are involved in the modulation of synaptic responses in the central nervous system and have been implicated in neuropsychological conditions that ...Metabotropic γ-aminobutyric acid receptors (GABA) are involved in the modulation of synaptic responses in the central nervous system and have been implicated in neuropsychological conditions that range from addiction to psychosis. GABA belongs to class C of the G-protein-coupled receptors, and its functional entity comprises an obligate heterodimer that is composed of the GB1 and GB2 subunits. Each subunit possesses an extracellular Venus flytrap domain, which is connected to a canonical seven-transmembrane domain. Here we present four cryo-electron microscopy structures of the human full-length GB1-GB2 heterodimer: one structure of its inactive apo state, two intermediate agonist-bound forms and an active form in which the heterodimer is bound to an agonist and a positive allosteric modulator. The structures reveal substantial differences, which shed light on the complex motions that underlie the unique activation mechanism of GABA. Our results show that agonist binding leads to the closure of the Venus flytrap domain of GB1, triggering a series of transitions, first rearranging and bringing the two transmembrane domains into close contact along transmembrane helix 6 and ultimately inducing conformational rearrangements in the GB2 transmembrane domain via a lever-like mechanism to initiate downstream signalling. This active state is stabilized by a positive allosteric modulator binding at the transmembrane dimerization interface. |
 External links External links |  Nature / Nature /  PubMed:32555460 / PubMed:32555460 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.63 - 6.3 Å |
| Structure data | EMDB-20822, PDB-6uo8: EMDB-20823, PDB-6uo9: EMDB-20824, PDB-6uoa: EMDB-21219, PDB-6vjm: |
| Chemicals | 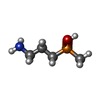 ChemComp-QD7:  ChemComp-NAG: 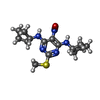 ChemComp-QDA: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / G protein-coupled receptor / GABA / GABAB / PAM / Neurotransmitter |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)