[English] 日本語
 Yorodumi
Yorodumi- EMDB-20504: 10 Angstrom structure of the asymmetric flagellar filament purifi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20504 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 10 Angstrom structure of the asymmetric flagellar filament purified from Leptospira biflexa Patoc WT cells resolved via subtomogram averaging | |||||||||||||||||||||||||||||||||
 Map data Map data | Masked full EM map resulting from subtomogram averaging in emClarity of flagellar filaments purified from wildtype Leptospira biflexa serovar Patoc strain Patoc 1. | |||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | bacterial flagella / FcpA / FcpB / FlaA / FlaB / Leptospira / STRUCTURAL PROTEIN | |||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||||||||||||||||||||||||||
| Biological species |  Leptospira biflexa serovar Patoc strain 'Patoc 1 (Paris)' (bacteria) / Leptospira biflexa serovar Patoc strain 'Patoc 1 (Paris)' (bacteria) /  Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) (bacteria) Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) (bacteria) | |||||||||||||||||||||||||||||||||
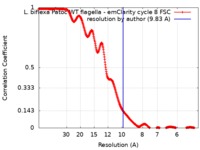
| Method | subtomogram averaging / cryo EM / Resolution: 9.83 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Gibson KH / Sindelar CV | |||||||||||||||||||||||||||||||||
| Funding support |  United States, Uruguay, 10 items United States, Uruguay, 10 items
| |||||||||||||||||||||||||||||||||
 Citation Citation | Journal: Front Cell Infect Microbiol / Year: 2018 Title: FcpB Is a Surface Filament Protein of the Endoflagellum Required for the Motility of the Spirochete . Authors: Elsio A Wunder / Leyla Slamti / David N Suwondo / Kimberley H Gibson / Zhiguo Shang / Charles V Sindelar / Felipe Trajtenberg / Alejandro Buschiazzo / Albert I Ko / Mathieu Picardeau /    Abstract: The spirochete endoflagellum is a unique motility apparatus among bacteria. Despite its critical importance for pathogenesis, the full composition of the flagellum remains to be determined. We have ...The spirochete endoflagellum is a unique motility apparatus among bacteria. Despite its critical importance for pathogenesis, the full composition of the flagellum remains to be determined. We have recently reported that FcpA is a novel flagellar protein and a major component of the sheath of the filament of the spirochete . By screening a library of random transposon mutants in the spirochete , we found a motility-deficient mutant harboring a disruption in a hypothetical gene of unknown function. Here, we show that this gene encodes a surface component of the endoflagellar filament and is required for typical hook- and spiral-shaped ends of the cell body, coiled structure of the endoflagella, and high velocity phenotype. We therefore named the gene for flagellar-coiling protein B. is conserved in all members of the genus, but not present in other organisms including other spirochetes. Complementation of the mutant restored the wild-type morphology and motility phenotypes. Immunoblotting with anti-FcpA and anti-FcpB antisera and cryo-electron microscopy of the filament indicated that FcpB assembled onto the surface of the sheath of the filament and mostly located on the outer (convex) side of the coiled filament. We provide evidence that FcpB, together with FcpA, are -specific novel components of the sheath of the filament, key determinants of the coiled and asymmetric structure of the endoflagella and are essential for high velocity. Defining the components of the endoflagella and their functions in these atypical bacteria should greatly enhance our understanding of the mechanisms by which these bacteria produce motility. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20504.map.gz emd_20504.map.gz | 25.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20504-v30.xml emd-20504-v30.xml emd-20504.xml emd-20504.xml | 40.4 KB 40.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20504_fsc.xml emd_20504_fsc.xml | 34.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_20504.png emd_20504.png | 97.9 KB | ||
| Filedesc metadata |  emd-20504.cif.gz emd-20504.cif.gz | 9.1 KB | ||
| Others |  emd_20504_additional_1.map.gz emd_20504_additional_1.map.gz emd_20504_additional_2.map.gz emd_20504_additional_2.map.gz emd_20504_additional_3.map.gz emd_20504_additional_3.map.gz emd_20504_additional_4.map.gz emd_20504_additional_4.map.gz emd_20504_half_map_1.map.gz emd_20504_half_map_1.map.gz emd_20504_half_map_2.map.gz emd_20504_half_map_2.map.gz | 25.5 MB 25.1 MB 25.5 MB 25.5 MB 25.1 MB 25.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20504 http://ftp.pdbj.org/pub/emdb/structures/EMD-20504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20504 | HTTPS FTP |
-Related structure data
| Related structure data |  6pwbMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20504.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20504.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked full EM map resulting from subtomogram averaging in emClarity of flagellar filaments purified from wildtype Leptospira biflexa serovar Patoc strain Patoc 1. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
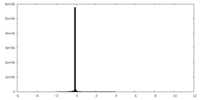
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.604 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
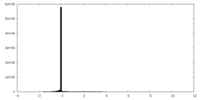
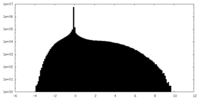
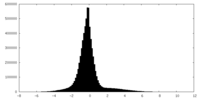
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Masked half map 1 resulting from subtomogram averaging...
| File | emd_20504_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked half map 1 resulting from subtomogram averaging in emClarity of flagellar filaments purified from wildtype Leptospira biflexa serovar Patoc strain Patoc 1. | ||||||||||||
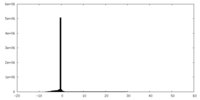
| Projections & Slices |
| ||||||||||||
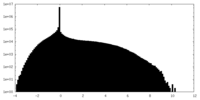
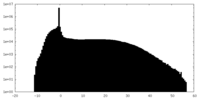
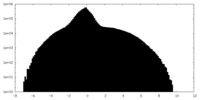
| Density Histograms |
-Additional map: Unmasked full EM map resulting from subtomogram averaging...
| File | emd_20504_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked full EM map resulting from subtomogram averaging in emClarity of flagellar filaments purified from wildtype Leptospira biflexa serovar Patoc strain Patoc 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
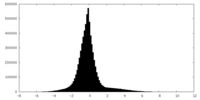
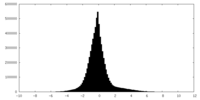
| Density Histograms |
-Additional map: Masked half map 2 resulting from subtomogram averaging...
| File | emd_20504_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked half map 2 resulting from subtomogram averaging in emClarity of flagellar filaments purified from wildtype Leptospira biflexa serovar Patoc strain Patoc 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
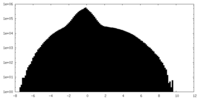
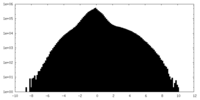
| Density Histograms |
-Additional map: Symmetrised map generated by rotational symmetrization of the...
| File | emd_20504_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetrised map generated by rotational symmetrization of the L. biflexa Patoc WT filament core resulting from subtomogram averaging. Map used to calculate helical parameters for FlaB core model. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unmasked-half map 1 resulting from subtomogram averaging in...
| File | emd_20504_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked-half map 1 resulting from subtomogram averaging in emClarity of flagellar filaments purified from wildtype Leptospira biflexa serovar Patoc strain Patoc 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unmasked-half map 2 resulting from subtomogram averaging in...
| File | emd_20504_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked-half map 2 resulting from subtomogram averaging in emClarity of flagellar filaments purified from wildtype Leptospira biflexa serovar Patoc strain Patoc 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Flagellar filament purified from the periplasm of the Spirochete ...
| Entire | Name: Flagellar filament purified from the periplasm of the Spirochete bacterium, Leptospira biflexa |
|---|---|
| Components |
|
-Supramolecule #1: Flagellar filament purified from the periplasm of the Spirochete ...
| Supramolecule | Name: Flagellar filament purified from the periplasm of the Spirochete bacterium, Leptospira biflexa type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Filaments were purified from Leptospira biflexa wild type cells. |
|---|---|
| Source (natural) | Organism:  Leptospira biflexa serovar Patoc strain 'Patoc 1 (Paris)' (bacteria) Leptospira biflexa serovar Patoc strain 'Patoc 1 (Paris)' (bacteria)Organelle: flagellar filament / Location in cell: periplasm |
-Macromolecule #1: Flagellin B1 (FlaB1)
| Macromolecule | Name: Flagellin B1 (FlaB1) / type: protein_or_peptide / ID: 1 / Number of copies: 84 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) (bacteria) Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) (bacteria)Strain: Patoc 1 / ATCC 23582 / Paris / Cell: bacteria |
| Molecular weight | Theoretical: 29.467033 KDa |
| Sequence | String: AAINSHRVLK FQNEEVSKNM EKLSSGMRIN RAGDDASGLA VSEKMRTQVN GLRQAERNTE DGMSLIQTTE GFLQESNDII QRIRTLAIQ SSNGIYTEED RQMIQVEVSQ LIDEVDRIAS QAEFNKMNLL QGDFARGSRA TSMWFHIGPN MHQRERVFIA T MTARSLNL ...String: AAINSHRVLK FQNEEVSKNM EKLSSGMRIN RAGDDASGLA VSEKMRTQVN GLRQAERNTE DGMSLIQTTE GFLQESNDII QRIRTLAIQ SSNGIYTEED RQMIQVEVSQ LIDEVDRIAS QAEFNKMNLL QGDFARGSRA TSMWFHIGPN MHQRERVFIA T MTARSLNL KGQSGELLSL STADKSNDAI GTLDAALTRI SKQRANLGAY FNRLEHAAKG LMNAYENTQA SESRIRDADM AE ETVAFTK NQILVQSGTA MLAQANVR UniProtKB: Flagellin |
-Macromolecule #2: Flagellar coiling protein A (FcpA)
| Macromolecule | Name: Flagellar coiling protein A (FcpA) / type: protein_or_peptide / ID: 2 / Number of copies: 47 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) (bacteria) Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) (bacteria)Strain: Patoc 1 / ATCC 23582 / Paris / Cell: bacteria |
| Molecular weight | Theoretical: 28.931002 KDa |
| Sequence | String: LTEDQKKKKK EIMEQESLWK NPDFKGYNKT FQELHQLSKT FANNQFRLAL SNYQSGVNTI MKNRDWVEQY RKEEAEKKRL DEKWYWQKV DRKAREERVV YREKMKAKQD ALNYFSKAIN HLDEIKNPDL RERPEFKRLL SDVYRSWIMA EYDLQNLPQT I PILELYIE ...String: LTEDQKKKKK EIMEQESLWK NPDFKGYNKT FQELHQLSKT FANNQFRLAL SNYQSGVNTI MKNRDWVEQY RKEEAEKKRL DEKWYWQKV DRKAREERVV YREKMKAKQD ALNYFSKAIN HLDEIKNPDL RERPEFKRLL SDVYRSWIMA EYDLQNLPQT I PILELYIE IDDNEKEYPA HKYLASAYSF EENMIKKTKG PDDMLFKYRY KKNVHLLRAT ELKYGKDSPE YKHIVNVIN UniProtKB: Uncharacterized protein |
-Macromolecule #3: Flagellar coiling protein B (FcpB)
| Macromolecule | Name: Flagellar coiling protein B (FcpB) / type: protein_or_peptide / ID: 3 / Number of copies: 32 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) (bacteria) Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) (bacteria)Strain: Patoc 1 / ATCC 23582 / Paris / Cell: bacteria |
| Molecular weight | Theoretical: 25.539666 KDa |
| Sequence | String: SGKSMADTEK ELDDNISEVN KRLRLHTVLF KMKVRTLPHK TVLYKGKPSA DGERCEAADK QEAQDNTCLH LEVFDFVGSE DGKSSKNLG AKFKKMELFF EGSNNADPDP RKEQPRNLTK IRTYIYQNNF LLEDKVISVI ADVAPNGEPA HNDKIELFYQ H DDYPVWGT ...String: SGKSMADTEK ELDDNISEVN KRLRLHTVLF KMKVRTLPHK TVLYKGKPSA DGERCEAADK QEAQDNTCLH LEVFDFVGSE DGKSSKNLG AKFKKMELFF EGSNNADPDP RKEQPRNLTK IRTYIYQNNF LLEDKVISVI ADVAPNGEPA HNDKIELFYQ H DDYPVWGT PETPSEKGVG KYILSNVENT KSNPIRNNFK KQFYFKNLDY FDKLFTKIFD YND UniProtKB: Uncharacterized protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 6.8 / Details: Tris-HCl or ddH20, pH6.8 with <1% sodium azide as a preservative. FIDUCIAL MARKERS: Prior to vitrification, 1 uL of 6x concentrated Gold Tracer beads (10 nm colloidal gold) were mixed with 2 uL of purified flagella. To each grid, 3 uL of this mixture were applied. |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 600 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER Details: Model 950 Solarus Advanced Plasma System manufactured by GATAN. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK III Details: 2 minute incubation time; blot time of 6-7.5 seconds; and blot offset of -2 mm. |
| Details | Leptospira biflexa serovar Patoc strain Patoc I wild type, fcpA, and fcpB mutant cells cultured in Ellinghausen-McCullough-Johnson-Harris liquid medium until they reached logarithmic phase at 30C. The cells were pelleted and the periplasmic flagellar filaments were purified as described in Wunder et al., 2016; Wunder et al., 2018. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-12 / Number grids imaged: 1 / Average exposure time: 1.2 sec. / Average electron dose: 1.7 e/Å2 Details: Tilt series acquisition. A total of ~35 tilt angles per tilt stack were acquired. The total dose was ~ 60 e/Angstrom2. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 19230 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 15000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | Eliminate minor clashes between FlaB1, FcpA, and FcpB | ||||||||
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient | ||||||||
| Output model |  PDB-6pwb: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)