[English] 日本語
 Yorodumi
Yorodumi- EMDB-20481: Structure of HIV cleaved synaptic complex (CSC) intasome bound wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20481 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of HIV cleaved synaptic complex (CSC) intasome bound with calcium | ||||||||||||
 Map data Map data | structure of HIV cleaved synaptic complex intasome in its apo form; sharpened | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | integrase / intasome / enzyme / transposition / VIRAL PROTEIN / VIRAL PROTEIN-DNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationRNA endonuclease activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA stem-loop binding / RNA-directed DNA polymerase activity / endonuclease activity / DNA recombination / symbiont entry into host cell / DNA binding ...RNA endonuclease activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA stem-loop binding / RNA-directed DNA polymerase activity / endonuclease activity / DNA recombination / symbiont entry into host cell / DNA binding / zinc ion binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||
 Authors Authors | Lyumkis D / Passos D | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural basis for strand-transfer inhibitor binding to HIV intasomes. Authors: Dario Oliveira Passos / Min Li / Ilona K Jóźwik / Xue Zhi Zhao / Diogo Santos-Martins / Renbin Yang / Steven J Smith / Youngmin Jeon / Stefano Forli / Stephen H Hughes / Terrence R Burke / ...Authors: Dario Oliveira Passos / Min Li / Ilona K Jóźwik / Xue Zhi Zhao / Diogo Santos-Martins / Renbin Yang / Steven J Smith / Youngmin Jeon / Stefano Forli / Stephen H Hughes / Terrence R Burke / Robert Craigie / Dmitry Lyumkis /  Abstract: The HIV intasome is a large nucleoprotein assembly that mediates the integration of a DNA copy of the viral genome into host chromatin. Intasomes are targeted by the latest generation of ...The HIV intasome is a large nucleoprotein assembly that mediates the integration of a DNA copy of the viral genome into host chromatin. Intasomes are targeted by the latest generation of antiretroviral drugs, integrase strand-transfer inhibitors (INSTIs). Challenges associated with lentiviral intasome biochemistry have hindered high-resolution structural studies of how INSTIs bind to their native drug target. Here, we present high-resolution cryo-electron microscopy structures of HIV intasomes bound to the latest generation of INSTIs. These structures highlight how small changes in the integrase active site can have notable implications for drug binding and design and provide mechanistic insights into why a leading INSTI retains efficacy against a broad spectrum of drug-resistant variants. The data have implications for expanding effective treatments available for HIV-infected individuals. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20481.map.gz emd_20481.map.gz | 200.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20481-v30.xml emd-20481-v30.xml emd-20481.xml emd-20481.xml | 27.2 KB 27.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20481.png emd_20481.png | 148.3 KB | ||
| Filedesc metadata |  emd-20481.cif.gz emd-20481.cif.gz | 7 KB | ||
| Others |  emd_20481_additional_1.map.gz emd_20481_additional_1.map.gz emd_20481_additional_2.map.gz emd_20481_additional_2.map.gz emd_20481_half_map_1.map.gz emd_20481_half_map_1.map.gz emd_20481_half_map_2.map.gz emd_20481_half_map_2.map.gz | 196.3 MB 11.5 MB 26.5 MB 26.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20481 http://ftp.pdbj.org/pub/emdb/structures/EMD-20481 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20481 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20481 | HTTPS FTP |
-Validation report
| Summary document |  emd_20481_validation.pdf.gz emd_20481_validation.pdf.gz | 715.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20481_full_validation.pdf.gz emd_20481_full_validation.pdf.gz | 714.7 KB | Display | |
| Data in XML |  emd_20481_validation.xml.gz emd_20481_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_20481_validation.cif.gz emd_20481_validation.cif.gz | 18.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20481 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20481 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20481 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20481 | HTTPS FTP |
-Related structure data
| Related structure data |  6putMC  6puwC  6puyC  6puzC  6v3kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20481.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20481.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | structure of HIV cleaved synaptic complex intasome in its apo form; sharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.79 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
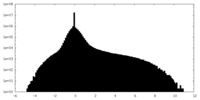
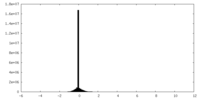
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Structure of HIV cleaved synaptic complex intasome in...
| File | emd_20481_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of HIV cleaved synaptic complex intasome in its apo form. Full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
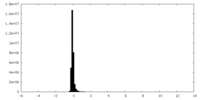
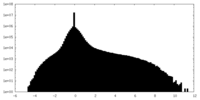
| Density Histograms |
-Additional map: local resolution map file
| File | emd_20481_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution map file | ||||||||||||
| Projections & Slices |
| ||||||||||||
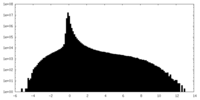
| Density Histograms |
-Half map: half map 2
| File | emd_20481_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
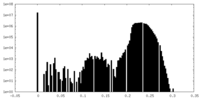
| Density Histograms |
-Half map: half map 1
| File | emd_20481_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
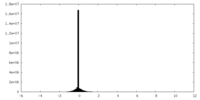
| Density Histograms |
- Sample components
Sample components
-Entire : Assembly of HIV integrase and viral DNA
| Entire | Name: Assembly of HIV integrase and viral DNA |
|---|---|
| Components |
|
-Supramolecule #1: Assembly of HIV integrase and viral DNA
| Supramolecule | Name: Assembly of HIV integrase and viral DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Chimeric Sso7d and HIV-1 integrase
| Macromolecule | Name: Chimeric Sso7d and HIV-1 integrase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 42.321258 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MATVKFKYKG EEKEVDISKI KKVWRVGKMI SFTYDEGGGK TGRGAVSEKD APKELLQMLE KQKKGGGGG GGGGGGFLDG IDKAQEEHEK YHSNWRAMAS DFNLPPVVAK EIVASCDKCQ LKGEAMHGQV DCSPGIWQLD C THLEGKVI ...String: MGSSHHHHHH SSGLVPRGSH MATVKFKYKG EEKEVDISKI KKVWRVGKMI SFTYDEGGGK TGRGAVSEKD APKELLQMLE KQKKGGGGG GGGGGGFLDG IDKAQEEHEK YHSNWRAMAS DFNLPPVVAK EIVASCDKCQ LKGEAMHGQV DCSPGIWQLD C THLEGKVI LVAVHVASGY IEAEVIPAET GQETAYFLLK LAGRWPVKTV HTDNGSNFTS TTVKAACWWA GIKQEFGIPY NP QSQGVIE SMNKELKKII GQVRDQAEHL KTAVQMAVFI HNFKRKGGIG GYSAGERIVD IIATDIQTKE LQKQITKIQN FRV YYRDSR DPVWKGPAKL LWKGEGAVVI QDNSDIKVVP RRKAKIIRDY GKQMAGDDCV ASRQDED UniProtKB: DNA-binding protein 7d, Gag-Pol polyprotein |
-Macromolecule #2: viral DNA transferred strand
| Macromolecule | Name: viral DNA transferred strand / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 7.773023 KDa |
| Sequence | String: (DA)(DG)(DC)(DG)(DT)(DG)(DG)(DG)(DC)(DG) (DG)(DG)(DA)(DA)(DA)(DA)(DT)(DC)(DT)(DC) (DT)(DA)(DG)(DC)(DA) |
-Macromolecule #3: viral DNA non-transferred strand
| Macromolecule | Name: viral DNA non-transferred strand / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 8.188271 KDa |
| Sequence | String: (DA)(DC)(DT)(DG)(DC)(DT)(DA)(DG)(DA)(DG) (DA)(DT)(DT)(DT)(DT)(DC)(DC)(DC)(DG)(DC) (DC)(DC)(DA)(DC)(DG)(DC)(DT) |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 154 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.2 Component:
| ||||||||||||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 4-80 / Number real images: 1989 / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 63291 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 37000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Correlation coefficient | ||||||||
| Output model |  PDB-6put: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)