[English] 日本語
 Yorodumi
Yorodumi- EMDB-20433: Kaposi's sarcoma-associated herpesvirus (KSHV), C1 penton vertex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20433 | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Kaposi's sarcoma-associated herpesvirus (KSHV), C1 penton vertex register, CATC-absent structure | |||||||||||||||||||||||||||||||||||||||||||||
 Map data Map data | None | |||||||||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | capsid / tegument / vertex / complex / VIRUS / VIRAL PROTEIN | |||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationT=16 icosahedral viral capsid / viral capsid assembly / viral process / viral capsid / host cell nucleus / structural molecule activity / DNA binding Similarity search - Function | |||||||||||||||||||||||||||||||||||||||||||||
| Biological species |   Human herpesvirus 8 / Human herpesvirus 8 /   Human gammaherpesvirus 8 Human gammaherpesvirus 8 | |||||||||||||||||||||||||||||||||||||||||||||
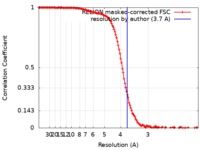
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Gong D / Dai X / Jih J / Liu YT / Bi GQ / Sun R / Zhou ZH | |||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  United States, United States,  China, 14 items China, 14 items
| |||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV. Authors: Danyang Gong / Xinghong Dai / Jonathan Jih / Yun-Tao Liu / Guo-Qiang Bi / Ren Sun / Z Hong Zhou /   Abstract: Assembly of Kaposi's sarcoma-associated herpesvirus (KSHV) begins at a bacteriophage-like portal complex that nucleates formation of an icosahedral capsid with capsid-associated tegument complexes ...Assembly of Kaposi's sarcoma-associated herpesvirus (KSHV) begins at a bacteriophage-like portal complex that nucleates formation of an icosahedral capsid with capsid-associated tegument complexes (CATCs) and facilitates translocation of an ∼150-kb dsDNA genome, followed by acquisition of a pleomorphic tegument and envelope. Because of deviation from icosahedral symmetry, KSHV portal and tegument structures have largely been obscured in previous studies. Using symmetry-relaxed cryo-EM, we determined the in situ structure of the KSHV portal and its interactions with surrounding capsid proteins, CATCs, and the terminal end of KSHV's dsDNA genome. Our atomic models of the portal and capsid/CATC, together with visualization of CATCs' variable occupancy and alternate orientation of CATC-interacting vertex triplexes, suggest a mechanism whereby the portal orchestrates procapsid formation and asymmetric long-range determination of CATC attachment during DNA packaging prior to pleomorphic tegumentation/envelopment. Structure-based mutageneses confirm that a triplex deep binding groove for CATCs is a hotspot that holds promise for antiviral development. | |||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20433.map.gz emd_20433.map.gz | 194.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20433-v30.xml emd-20433-v30.xml emd-20433.xml emd-20433.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20433_fsc.xml emd_20433_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_20433.png emd_20433.png | 241.8 KB | ||
| Masks |  emd_20433_msk_1.map emd_20433_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20433.cif.gz emd-20433.cif.gz | 7.4 KB | ||
| Others |  emd_20433_half_map_1.map.gz emd_20433_half_map_1.map.gz emd_20433_half_map_2.map.gz emd_20433_half_map_2.map.gz | 169 MB 169 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20433 http://ftp.pdbj.org/pub/emdb/structures/EMD-20433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20433 | HTTPS FTP |
-Related structure data
| Related structure data |  6ppdMC  6ppbC  6pphC  6ppiC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20433.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20433.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20433_msk_1.map emd_20433_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: None
| File | emd_20433_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: None
| File | emd_20433_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human gammaherpesvirus 8
| Entire | Name:   Human gammaherpesvirus 8 Human gammaherpesvirus 8 |
|---|---|
| Components |
|
-Supramolecule #1: Human gammaherpesvirus 8
| Supramolecule | Name: Human gammaherpesvirus 8 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 37296 / Sci species name: Human gammaherpesvirus 8 / Sci species strain: BAC16 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 1250.0 Å / T number (triangulation number): 16 |
-Macromolecule #1: Small capsomere-interacting protein
| Macromolecule | Name: Small capsomere-interacting protein / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 8 / Strain: GK18 Human herpesvirus 8 / Strain: GK18 |
| Molecular weight | Theoretical: 18.597824 KDa |
| Sequence | String: MSNFKVRDPV IQERLDHDYA HHPLVARMNT LDQGNMSQAE YLVQKRHYLV FLIAHHYYEA YLRRMGGIQR RDHLQTLRDQ KPRERADRV SAASAYDAGT FTVPSRPGPA SGTTPGGQDS LGVSGSSITT LSSGPHSLSP ASDILTTLSS TTETAAPAVA D ARKPPSGK KK UniProtKB: Small capsomere-interacting protein |
-Macromolecule #2: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 8 / Strain: GK18 Human herpesvirus 8 / Strain: GK18 |
| Molecular weight | Theoretical: 153.574188 KDa |
| Sequence | String: MEATLEQRPF PYLATEANLL TQIKESAADG LFKSFQLLLG KDAREGSVRF EALLGVYTNV VEFVKFLETA LAAACVNTEF KDLRRMIDG KIQFKISMPT IAHGDGRRPN KQRQYIVMKA CNKHHIGAEI ELAAADIELL FAEKETPLDF TEYAGAIKTI T SALQFGMD ...String: MEATLEQRPF PYLATEANLL TQIKESAADG LFKSFQLLLG KDAREGSVRF EALLGVYTNV VEFVKFLETA LAAACVNTEF KDLRRMIDG KIQFKISMPT IAHGDGRRPN KQRQYIVMKA CNKHHIGAEI ELAAADIELL FAEKETPLDF TEYAGAIKTI T SALQFGMD ALERGLVDTV LAVKLRHAPP VFILKTLGDP VYSERGLKKA VKSDMVSMFK AHLIEHSFFL DKAELMTRGK QY VLTMLSD MLAAVCEDTV FKGVSTYTTA SGQQVAGVLE TTDSVMRRLM NLLGQVESAM SGPAAYASYV VRGANLVTAV SYG RAMRNF EQFMARIVDH PNALPSVEGD KAALADGHDE IQRTRIAASL VKIGDKFVAI ESLQRMYNET QFPCPLNRRI QYTY FFPVG LHLPVPRYST SVSVRGVESP AIQSTETWVV NKNNVPLCFG YQNALKSICH PRMHNPTQSA QALNQAFPDP DGGHG YGLR YEQTPNMNLF RTFHQYYMGK NVAFVPDVAQ KALVTTEDLL HPTSHRLLRL EVHPFFDFFV HPCPGARGSY RATHRT MVG NIPQPLAPRE FQESRGAQFD AVTNMTHVID QLTIDVIQET AFDPAYPLFC YVIEAMIHGQ EEKFVMNMPL IALVIQT YW VNSGKLAFVN SYHMVRFICT HMGNGSIPKE AHGHYRKILG ELIALEQALL KLAGHETVGR TPITHLVSAL LDPHLLPP F AYHDVFTDLM QKSSRQPIIK IGDQNYDNPQ NRATFINLRG RMEDLVNNLV NIYQTRVNED HDERHVLDVA PLDENDYNP VLEKLFYYVL MPVCSNGHMC GMGVDYQNVA LTLTYNGPVF ADVVNAQDDI LLHLENGTLK DILQAGDIRP TVDMIRVLCT SFLTCPFVT QAARVITKRD PAQSFATHEY GKDVAQTVLV NGFGAFAVAD RSREAAETMF YPVPFNKLYA DPLVAATLHP L LANYVTRL PNQRNAVVFN VPSNLMAEYE EWHKSPVAAY AASCQATPGA ISAMVSMHQK LSAPSFICQA KHRMHPGFAM TV VRTDEVL AEHILYCSRA STSMFVGLPS VVRREVRSDA VTFEITHEIA SLHTALGYSS VIAPAHVAAI TTDMGVHCQD LFM IFPGDA YQDRQLHDYI KMKAGVQTGP PGNRMDHVGY AAGVPRCENL PGLSHGQLAT CEIIPTPVTS DVAYFQTPSN PRGR AACVV SCDAYSNESA ERLLYDHSIP DPAYECRSTN NPWASQRGSL GDVLYNITFR QTALPGMYSP CRQFFHKEDI MRYNR GLYT LVNEYSARLA GAPATSTTDL QYVVVNGTDV FLDQPCHMLQ EAYPTLAASH RVMLDEYMSN KQTHAPVHMG QYLIEE VAP MKRLLKLGNK VVY UniProtKB: ORF25 |
-Macromolecule #3: Triplex capsid protein 1
| Macromolecule | Name: Triplex capsid protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 8 / Strain: GK18 Human herpesvirus 8 / Strain: GK18 |
| Molecular weight | Theoretical: 36.37484 KDa |
| Sequence | String: MKVQAENAAR LGRQVLGLLP PPTHRVSLTR GPEFARGVRD LLSKYAASTR PTVGSLHEAL RQAPFRQPTY GDFLVYSQTF SPQEPLGTF LFSFKQEDNG SSMDMLLTPT SLFMLSGMEA AKAPQTHKVA GVWYGSGSGL ADFIPNLSEL MDTGEFHTLL T PVGPMVQS ...String: MKVQAENAAR LGRQVLGLLP PPTHRVSLTR GPEFARGVRD LLSKYAASTR PTVGSLHEAL RQAPFRQPTY GDFLVYSQTF SPQEPLGTF LFSFKQEDNG SSMDMLLTPT SLFMLSGMEA AKAPQTHKVA GVWYGSGSGL ADFIPNLSEL MDTGEFHTLL T PVGPMVQS VHSTFVTKVT SAMKGVGLAR DEPRAHVGLT LPCDMLVDLD ESCPMVQRRE PAGLNVTIYA SLVYLRVNQR PS MALTFFQ SGKGFAEVVA MIKDHFTDVI RTKYIQLRHE LYINRLVFGA VCTLGTVPFD SHPVHQSLNV KGTSLPVLVF ANF EAACGP WTVFL UniProtKB: Core gene UL38 family protein |
-Macromolecule #4: Triplex capsid protein 2
| Macromolecule | Name: Triplex capsid protein 2 / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 8 / Strain: GK18 Human herpesvirus 8 / Strain: GK18 |
| Molecular weight | Theoretical: 34.278473 KDa |
| Sequence | String: MALDKSIVVN LTSRLFADEL AALQSKIGSV LPLGDCHRLQ NIQALGLGCV CSRETSPDYI QIMQYLSKCT LAVLEEVRPD SLRLTRMDP SDNLQIKNVY APFFQWDSNT QLAVLPPLFS RKDSTIVLES NGFDIVFPMV VPQQLGHAIL QQLLVYHIYS K ISAGAPGD ...String: MALDKSIVVN LTSRLFADEL AALQSKIGSV LPLGDCHRLQ NIQALGLGCV CSRETSPDYI QIMQYLSKCT LAVLEEVRPD SLRLTRMDP SDNLQIKNVY APFFQWDSNT QLAVLPPLFS RKDSTIVLES NGFDIVFPMV VPQQLGHAIL QQLLVYHIYS K ISAGAPGD VNMAELDLYT TNVSFMGRTY RLDVDNTDPR TALRVLDDLS MYLCILSALV PRGCLRLLTA LVRHDRHPLT EV FEGVVPD EVTRIDLDQL SVPDDITRMR VMFSYLQSLS SIFNLGPRLH VYAYSAETLA ASCWYSPR UniProtKB: ORF26 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 298 K / Instrument: HOMEMADE PLUNGER Details: The sample was manually blotted and frozen with a homemade plunger.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Max: 79.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Dimensions - Width: 1440 pixel / Digitization - Dimensions - Height: 1440 pixel / Number real images: 8007 / Average exposure time: 13.0 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 24271 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 14000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 177.4 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6ppd: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)