+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ha3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | ELONGATION FACTOR TU IN COMPLEX WITH aurodox | ||||||
 Components Components | ELONGATION FACTOR TU | ||||||
 Keywords Keywords | TRANSLATION / GTPASE / AURODOX / N-METHYL-KIRROMYCIN / ANTIBIOTIC / RIBOSOME | ||||||
| Function / homology |  Function and homology information Function and homology informationprotein-synthesizing GTPase / translation elongation factor activity / GTPase activity / GTP binding / RNA binding / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |   THERMUS THERMOPHILUS (bacteria) THERMUS THERMOPHILUS (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Vogeley, L. / Palm, G.J. / Mesters, J.R. / Hilgenfeld, R. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2001 Journal: J.Biol.Chem. / Year: 2001Title: Conformational Change of Elongation Factor TU Induced by Antibiotic Binding: Crystal Structure of the Complex between EF-TU:Gdp and Aurodox Authors: Vogeley, L. / Palm, G.J. / Mesters, J.R. / Hilgenfeld, R. #1:  Journal: The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions Journal: The Ribosome: Structure, Function, Antibiotics, and Cellular InteractionsYear: 2000 Title: Insights Into the Gtpase Mechanism of EF-TU from Structural Studies Authors: Hilgenfeld, R. / Mesters, J. / Hogg, T. #3: Journal: Nature / Year: 1993 Title: Crystal Structure of Active Elongation Factor TU Reveals Major Domain Rearrangements Authors: Berchtold, H. / Reshetnikova, L. / Reiser, C.O.A. / Schirmer, N.K. / Sprinzl, M. / Hilgenfeld, R. #4:  Journal: J.Cryst.Growth / Year: 1992 Journal: J.Cryst.Growth / Year: 1992Title: Crystals of Intact Elongation Factor TU from Thermus Thermophilus Diffracting to 1.45 Angstrom Resolution Authors: Reshetnikova, L. / Schirmer, N.K. / Reiser, C.O.A. / Berchtold, H. / Storm, R. / Hilgenfeld, R. / Sprinzl, M. | ||||||
| History |
| ||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "D" IN EACH CHAIN ON SHEET RECORDS BELOW ... SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "D" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 6-STRANDED BARREL THIS IS REPRESENTED BY A 7-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ha3.cif.gz 1ha3.cif.gz | 182.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ha3.ent.gz pdb1ha3.ent.gz | 142.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ha3.json.gz 1ha3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ha/1ha3 https://data.pdbj.org/pub/pdb/validation_reports/ha/1ha3 ftp://data.pdbj.org/pub/pdb/validation_reports/ha/1ha3 ftp://data.pdbj.org/pub/pdb/validation_reports/ha/1ha3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1exmS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (0.981974, -0.188989, -0.003066), Vector: |
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 44709.887 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: TERNARY COMPLEX WITH GUANOSINE DIPHOSPHATE AND AURODOX Source: (gene. exp.)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 / Gene: TUFB / Plasmid: PEFTU10 / Production host: THERMUS THERMOPHILUS (bacteria) / Strain: HB8 / Gene: TUFB / Plasmid: PEFTU10 / Production host:  |
|---|
-Non-polymers , 5 types, 606 molecules 

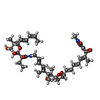






| #2: Chemical | | #3: Chemical | #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.8 Å3/Da / Density % sol: 55 % Description: DOMAIN 1 AND 2/3 HAD TO BE USED AS SEPARATE SEARCH MODELS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 8 Details: HANGING DROP METHOD (PROTEIN SOLUTION:WELL 1:1). PROTEIN CONCENTRATION 10 MG/ML. WELL: 50 MM TRIS, 200 MM NAOAC, 23-25% PEG4000, PH 8.0 MOLAR RATIO EF-TU:GDP:AURODOX = 1:1:5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 19 ℃ / pH: 7 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 105 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X13 / Wavelength: 0.913 / Beamline: X13 / Wavelength: 0.913 |
| Detector | Type: X-RAY RESEARCH, HAMBURG, GERMANY / Detector: CCD / Date: Sep 15, 2000 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.913 Å / Relative weight: 1 |
| Reflection | Resolution: 2→60 Å / Num. obs: 67199 / % possible obs: 99.8 % / Observed criterion σ(I): -3 / Redundancy: 4.3 % / Biso Wilson estimate: 24.7 Å2 / Rsym value: 0.067 / Net I/σ(I): 18.2 |
| Reflection shell | Resolution: 2→2.03 Å / Redundancy: 2.8 % / Mean I/σ(I) obs: 3.9 / Rsym value: 0.263 / % possible all: 98.7 |
| Reflection | *PLUS Num. measured all: 287768 / Rmerge(I) obs: 0.067 |
| Reflection shell | *PLUS % possible obs: 98.7 % / Num. unique obs: 3322 / Num. measured obs: 9419 / Rmerge(I) obs: 0.263 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1EXM Resolution: 2→20 Å / Rfactor Rfree error: 0.004 / Data cutoff high absF: 1000000 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 51.18 Å2 / ksol: 0.36 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 30.4 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2→2.07 Å / Rfactor Rfree error: 0.016 / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller














 PDBj
PDBj












