[English] 日本語
 Yorodumi
Yorodumi- PDB-1gte: DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1gte | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | |||||||||
 Components Components | DIHYDROPYRIMIDINE DEHYDROGENASE | |||||||||
 Keywords Keywords | OXIDOREDUCTASE / ELECTRON TRANSFER / FLAVIN / IRON-SULFUR CLUSTERS / PYRIMIDINE CATABOLISM / 5-FLUOROURACIL DEGRADATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationdihydropyrimidine dehydrogenase (NADP+) / uracil binding / thymidine catabolic process / dihydropyrimidine dehydrogenase (NADP+) activity / beta-alanine biosynthetic process / uracil catabolic process / thymine catabolic process / FMN binding / NADP binding / flavin adenine dinucleotide binding ...dihydropyrimidine dehydrogenase (NADP+) / uracil binding / thymidine catabolic process / dihydropyrimidine dehydrogenase (NADP+) activity / beta-alanine biosynthetic process / uracil catabolic process / thymine catabolic process / FMN binding / NADP binding / flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding / protein homodimerization activity / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.65 Å MOLECULAR REPLACEMENT / Resolution: 1.65 Å | |||||||||
 Authors Authors | Dobritzsch, D. / Ricagno, S. / Schneider, G. / Schnackerz, K.D. / Lindqvist, Y. | |||||||||
 Citation Citation |  Journal: J. Biol. Chem. / Year: 2002 Journal: J. Biol. Chem. / Year: 2002Title: Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer. Authors: Dobritzsch, D. / Ricagno, S. / Schneider, G. / Schnackerz, K.D. / Lindqvist, Y. #1: Journal: Acta Crystallogr.,Sect.D / Year: 2001 Title: Crystallization and Preliminary X-Ray Study of Pig Liver Dihydropyrimidine Dehydrogenase Authors: Dobritzsch, D. / Persson, K. / Schneider, G. / Lindqvist, Y. | |||||||||
| History |
| |||||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AF", "BF", "CF" AND "DF" IN EACH CHAIN ... SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AF", "BF", "CF" AND "DF" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 8-STRANDED BARREL, THIS IS REPRESENTED BY A 9-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1gte.cif.gz 1gte.cif.gz | 895.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1gte.ent.gz pdb1gte.ent.gz | 720.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1gte.json.gz 1gte.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gt/1gte https://data.pdbj.org/pub/pdb/validation_reports/gt/1gte ftp://data.pdbj.org/pub/pdb/validation_reports/gt/1gte ftp://data.pdbj.org/pub/pdb/validation_reports/gt/1gte | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||
| 2 | 
| ||||||||||||||||
| Unit cell |
| ||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein | Mass: 111603.344 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q28943, dihydropyrimidine dehydrogenase (NADP+) |
|---|
-Non-polymers , 5 types, 4876 molecules 


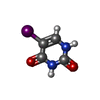





| #2: Chemical | ChemComp-SF4 / #3: Chemical | ChemComp-FMN / #4: Chemical | ChemComp-FAD / #5: Chemical | ChemComp-IUR / #6: Water | ChemComp-HOH / | |
|---|
-Details
| Compound details | INVOLVED IN THE CATABOLISM OF URACIL AND THYMIDINE LEADING TO THE FORMATION OF BETA-ALANINE. ...INVOLVED IN THE CATABOLISM |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.35 Å3/Da / Density % sol: 38.4 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 4.7 Details: 100 MM SODIUM CITRATE PH 4.7, 16-20 % POLYETHYLENE GLYCOL 6000, 1 MM DTT, 1 MM 5-IODOURACIL | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 293 K / Method: vapor diffusion / Details: Dobritzsch, D., (2001) Acta Crystallogr., 57, 153. | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: BW7B / Wavelength: 0.8424 / Beamline: BW7B / Wavelength: 0.8424 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Feb 26, 2000 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.8424 Å / Relative weight: 1 |
| Reflection | Resolution: 1.65→30 Å / Num. obs: 489160 / % possible obs: 98.1 % / Redundancy: 3.2 % / Biso Wilson estimate: 17.3 Å2 / Rmerge(I) obs: 0.047 / Net I/σ(I): 15 |
| Reflection shell | Resolution: 1.65→1.74 Å / Redundancy: 2.7 % / Rmerge(I) obs: 0.238 / Mean I/σ(I) obs: 4.3 / % possible all: 94.3 |
| Reflection | *PLUS Lowest resolution: 25 Å / Num. obs: 481684 |
| Reflection shell | *PLUS % possible obs: 94.3 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: DIHYDROPYRIMIDINE DEHYDROGENASE, NON-LIGANDED FORM Resolution: 1.65→24.98 Å / Rfactor Rfree error: 0.002 / Data cutoff high absF: 3440087 / Isotropic thermal model: OVERALL / Cross valid method: THROUGHOUT / σ(F): 0 Details: THE 5 C-TERMINAL RESIDUES AND A FEW LOOP-RESIDUES WERE NOT SEEN IN THE DENSITY DUE TO DISORDER
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 51.8663 Å2 / ksol: 0.394487 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.65→24.98 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.65→1.73 Å / Rfactor Rfree error: 0.007 / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.225 |
 Movie
Movie Controller
Controller
















 PDBj
PDBj













