[English] 日本語
 Yorodumi
Yorodumi- PDB-1foj: BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1foj | ||||||
|---|---|---|---|---|---|---|---|
| Title | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 7-NITROINDAZOLE-2-CARBOXAMIDINE (H4B PRESENT) | ||||||
 Components Components | NITRIC-OXIDE SYNTHASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE / alpha-beta fold / NITRIC OXIDE SYNTHASE | ||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to laminar fluid shear stress / negative regulation of leukocyte cell-cell adhesion / nitric oxide mediated signal transduction / nitric-oxide synthase (NADPH) / nitric-oxide synthase activity / L-arginine catabolic process / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of blood pressure / response to hormone / nitric oxide biosynthetic process ...cellular response to laminar fluid shear stress / negative regulation of leukocyte cell-cell adhesion / nitric oxide mediated signal transduction / nitric-oxide synthase (NADPH) / nitric-oxide synthase activity / L-arginine catabolic process / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of blood pressure / response to hormone / nitric oxide biosynthetic process / mitochondrion organization / caveola / blood coagulation / FMN binding / flavin adenine dinucleotide binding / NADP binding / response to lipopolysaccharide / cytoskeleton / calmodulin binding / heme binding / Golgi apparatus / metal ion binding / nucleus / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.1 Å SYNCHROTRON / Resolution: 2.1 Å | ||||||
 Authors Authors | Raman, C.S. / Li, H. / Martasek, P. / Masters, B.S. / Poulos, T.L. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2001 Journal: Biochemistry / Year: 2001Title: Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism Authors: Raman, C.S. / Li, H. / Martasek, P. / Southan, G. / Masters, B.S. / Poulos, T.L. #1:  Journal: Cell(Cambridge,Mass.) / Year: 1998 Journal: Cell(Cambridge,Mass.) / Year: 1998Title: Crystal structure of constitutive endothelial nitric oxide synthase: A paradigm for pterin function involving a novel metal center Authors: Raman, C.S. / Li, H. / Martasek, P. / Kral, V. / Masters, B.S. / Poulos, T.L. #2:  Journal: Science / Year: 1998 Journal: Science / Year: 1998Title: Structure of nitric oxide synthase oxygenase dimer with pterin and substrate Authors: Crane, B.R. / Arvai, A.S. / Ghosh, D.K. / Wu, C. / Getzoff, E.D. / Stuehr, D.J. / Tainer, J.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1foj.cif.gz 1foj.cif.gz | 190.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1foj.ent.gz pdb1foj.ent.gz | 148.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1foj.json.gz 1foj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1foj_validation.pdf.gz 1foj_validation.pdf.gz | 1.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1foj_full_validation.pdf.gz 1foj_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  1foj_validation.xml.gz 1foj_validation.xml.gz | 37.3 KB | Display | |
| Data in CIF |  1foj_validation.cif.gz 1foj_validation.cif.gz | 52.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fo/1foj https://data.pdbj.org/pub/pdb/validation_reports/fo/1foj ftp://data.pdbj.org/pub/pdb/validation_reports/fo/1foj ftp://data.pdbj.org/pub/pdb/validation_reports/fo/1foj | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 49710.105 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: CACODYLATE (RESIDUE CAC) BINDS TO SG CYS 384 OF BOTH CHAINS Source: (gene. exp.)   |
|---|
-Non-polymers , 7 types, 385 molecules 



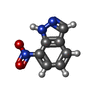
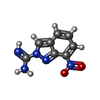







| #2: Chemical | | #3: Chemical | ChemComp-ACT / #4: Chemical | ChemComp-ZN / | #5: Chemical | #6: Chemical | #7: Chemical | #8: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.43 Å3/Da / Density % sol: 49.42 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 280 K / Method: vapor diffusion, sitting drop / pH: 6.5 Details: PEG 4000, cacodylate, magnesium acetate, pH 6.5, VAPOR DIFFUSION, SITTING DROP at 280K | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Details: Raman, C.S., (1998) Cell (Cambridge,Mass.), 95, 939. | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-1 / Wavelength: 0.78 / Beamline: BL9-1 / Wavelength: 0.78 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Feb 20, 1999 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.78 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→20 Å / Num. obs: 54878 / % possible obs: 95.2 % / Observed criterion σ(I): -3 / Redundancy: 2.6 % / Biso Wilson estimate: 30.8 Å2 / Rmerge(I) obs: 0.058 / Net I/σ(I): 8.4 |
| Reflection shell | Resolution: 2.1→2.14 Å / Redundancy: 2.2 % / Rmerge(I) obs: 0.599 / Num. unique all: 54878 / % possible all: 96.5 |
| Reflection | *PLUS Highest resolution: 2.1 Å / Num. measured all: 141597 |
| Reflection shell | *PLUS % possible obs: 96.5 % / Rmerge(I) obs: 0.761 / Mean I/σ(I) obs: 1.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.1→19.85 Å / Rfactor Rfree error: 0.006 / Data cutoff high absF: 478293.35 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 56.08 Å2 / ksol: 0.367 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 45.1 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→19.85 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.1→2.18 Å / Rfactor Rfree error: 0.032 / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 0.9 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS σ(F): 0 / % reflection Rfree: 4.4 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 45.1 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.43 / % reflection Rfree: 4.2 % / Rfactor Rwork: 0.423 |
 Movie
Movie Controller
Controller



























 PDBj
PDBj










