[English] 日本語
 Yorodumi
Yorodumi- PDB-1b2l: ALCOHOL DEHYDROGENASE FROM DROSOPHILA LEBANONENSIS: TERNARY COMPL... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1b2l | ||||||
|---|---|---|---|---|---|---|---|
| Title | ALCOHOL DEHYDROGENASE FROM DROSOPHILA LEBANONENSIS: TERNARY COMPLEX WITH NAD-CYCLOHEXANONE | ||||||
 Components Components | ALCOHOL DEHYDROGENASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE / DETOXIFICATION / METABOLISM / ALCOHOL DEHYDROGENASE / DROSOPHILA LEBANONENSIS / SHORT-CHAIN DEHYDROGENASES/REDUCTASES / TERNARY COMPLEX / NAD- CYCLOHEXANONE ADDUCT | ||||||
| Function / homology |  Function and homology information Function and homology informationalcohol metabolic process / alcohol dehydrogenase (NAD+) activity / alcohol dehydrogenase / identical protein binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  Scaptodrosophila lebanonensis (fry) Scaptodrosophila lebanonensis (fry) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å MOLECULAR REPLACEMENT / Resolution: 1.6 Å | ||||||
 Authors Authors | Benach, J. / Atrian, S. / Gonzalez-Duarte, R. / Ladenstein, R. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography. Authors: Benach, J. / Atrian, S. / Gonzalez-Duarte, R. / Ladenstein, R. #1:  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: The Refined Crystal Structure of Drosophila Lebanonensis Alcohol Dehydrogenase at 1.9 A Resolution Authors: Benach, J. / Atrian, S. / Gonzalez-Duarte, R. / Ladenstein, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1b2l.cif.gz 1b2l.cif.gz | 71.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1b2l.ent.gz pdb1b2l.ent.gz | 51 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1b2l.json.gz 1b2l.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b2/1b2l https://data.pdbj.org/pub/pdb/validation_reports/b2/1b2l ftp://data.pdbj.org/pub/pdb/validation_reports/b2/1b2l ftp://data.pdbj.org/pub/pdb/validation_reports/b2/1b2l | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1b14C  1b15C  1b16C  1a4uS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 27823.973 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: CYCLOHEXANONE / Source: (natural)  Scaptodrosophila lebanonensis (fry) / References: UniProt: P10807, alcohol dehydrogenase Scaptodrosophila lebanonensis (fry) / References: UniProt: P10807, alcohol dehydrogenase |
|---|
-Non-polymers , 5 types, 236 molecules 
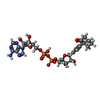







| #2: Chemical | ChemComp-CA / |
|---|---|
| #3: Chemical | ChemComp-NDC / |
| #4: Chemical | ChemComp-CYH / |
| #5: Chemical | ChemComp-DTT / |
| #6: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.56 Å3/Da / Density % sol: 51.99 % | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / pH: 8 Details: PROTEIN WAS CRYSTALLIZED FROM 20% PEG 2000, 0.2 M CACL2, 0.1 M TRIS-HCL, PH= 8.0, 1MM NAD+, 1% CYCLOHEXANONE, 277 K. | ||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 8.6 / Method: vapor diffusion, sitting drop / Details: Ladenstein, R., (1995) Acta Crystallog., D51, 69. | ||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  MAX II MAX II  / Beamline: I711 / Wavelength: 0.996 / Beamline: I711 / Wavelength: 0.996 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jun 1, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.996 Å / Relative weight: 1 |
| Reflection | Resolution: 1.57→20 Å / Num. obs: 40649 / % possible obs: 99 % / Observed criterion σ(I): -3 / Redundancy: 3 % / Biso Wilson estimate: 15.1 Å2 / Rmerge(I) obs: 0.057 / Net I/σ(I): 3 |
| Reflection shell | Resolution: 1.57→1.63 Å / Redundancy: 3 % / Rmerge(I) obs: 0.113 / Mean I/σ(I) obs: 3 / % possible all: 91 |
| Reflection | *PLUS Num. measured all: 565296 |
| Reflection shell | *PLUS % possible obs: 91 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1A4U Resolution: 1.6→8 Å / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.16 / ESU R Free: 0.18
| ||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 11.3 Å2 | ||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.6→8 Å
| ||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.6 Å / σ(F): 0 / % reflection Rfree: 5 % / Rfactor obs: 0.19 / Rfactor Rwork: 0.19 | ||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 11.3 Å2 | ||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller












 PDBj
PDBj












