[English] 日本語
 Yorodumi
Yorodumi- EMDB-14879: Mre11-Rad50-Nbs1 complex (Chaetomium thermophilum) lower coiled-coils -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mre11-Rad50-Nbs1 complex (Chaetomium thermophilum) lower coiled-coils | ||||||||||||
 Map data Map data | Lower CC, main map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | DNA repair / complex / ATPase / coiled-coils / HYDROLASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationMre11 complex / 3'-5'-DNA exonuclease activity / DNA endonuclease activity / meiotic cell cycle / double-strand break repair / manganese ion binding / chromosome, telomeric region / Hydrolases; Acting on ester bonds Similarity search - Function | ||||||||||||
| Biological species |  Chaetomium thermophilum (fungus) / Chaetomium thermophilum (fungus) /  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) | ||||||||||||
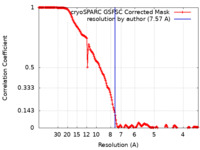
| Method | single particle reconstruction / cryo EM / Resolution: 7.57 Å | ||||||||||||
 Authors Authors | Stakyte K / Rotheneder M / Bartho JD / Lammens K / Hopfner KP | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions. Authors: Matthias Rotheneder / Kristina Stakyte / Erik van de Logt / Joseph D Bartho / Katja Lammens / Yilan Fan / Aaron Alt / Brigitte Kessler / Christophe Jung / Wynand P Roos / Barbara ...Authors: Matthias Rotheneder / Kristina Stakyte / Erik van de Logt / Joseph D Bartho / Katja Lammens / Yilan Fan / Aaron Alt / Brigitte Kessler / Christophe Jung / Wynand P Roos / Barbara Steigenberger / Karl-Peter Hopfner /  Abstract: The DNA double-strand break repair complex Mre11-Rad50-Nbs1 (MRN) detects and nucleolytically processes DNA ends, activates the ATM kinase, and tethers DNA at break sites. How MRN can act both as ...The DNA double-strand break repair complex Mre11-Rad50-Nbs1 (MRN) detects and nucleolytically processes DNA ends, activates the ATM kinase, and tethers DNA at break sites. How MRN can act both as nuclease and scaffold protein is not well understood. The cryo-EM structure of MRN from Chaetomium thermophilum reveals a 2:2:1 complex with a single Nbs1 wrapping around the autoinhibited Mre11 nuclease dimer. MRN has two DNA-binding modes, one ATP-dependent mode for loading onto DNA ends and one ATP-independent mode through Mre11's C terminus, suggesting how it may interact with DSBs and intact DNA. MRNs two 60-nm-long coiled-coil domains form a linear rod structure, the apex of which is assembled by the two joined zinc-hook motifs. Apices from two MRN complexes can further dimerize, forming 120-nm spanning MRN-MRN structures. Our results illustrate the architecture of MRN and suggest how it mechanistically integrates catalytic and tethering functions. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14879.map.gz emd_14879.map.gz | 450 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14879-v30.xml emd-14879-v30.xml emd-14879.xml emd-14879.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14879_fsc.xml emd_14879_fsc.xml | 18.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_14879.png emd_14879.png | 23.6 KB | ||
| Masks |  emd_14879_msk_1.map emd_14879_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14879.cif.gz emd-14879.cif.gz | 7.2 KB | ||
| Others |  emd_14879_half_map_1.map.gz emd_14879_half_map_1.map.gz emd_14879_half_map_2.map.gz emd_14879_half_map_2.map.gz | 441.8 MB 441.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14879 http://ftp.pdbj.org/pub/emdb/structures/EMD-14879 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14879 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14879 | HTTPS FTP |
-Validation report
| Summary document |  emd_14879_validation.pdf.gz emd_14879_validation.pdf.gz | 754.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14879_full_validation.pdf.gz emd_14879_full_validation.pdf.gz | 754.4 KB | Display | |
| Data in XML |  emd_14879_validation.xml.gz emd_14879_validation.xml.gz | 24.9 KB | Display | |
| Data in CIF |  emd_14879_validation.cif.gz emd_14879_validation.cif.gz | 32.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14879 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14879 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14879 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14879 | HTTPS FTP |
-Related structure data
| Related structure data |  7zqyC  7zr1C  8bahC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14879.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14879.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Lower CC, main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.8003 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14879_msk_1.map emd_14879_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Lower CC, half map B
| File | emd_14879_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Lower CC, half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Lower CC, half map A
| File | emd_14879_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Lower CC, half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS
| Entire | Name: Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS |
|---|---|
| Components |
|
-Supramolecule #1: Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS
| Supramolecule | Name: Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
-Macromolecule #1: Double-strand break repair protein
| Macromolecule | Name: Double-strand break repair protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MPQTAGPDTI RILVSTDNHV GYEERDPIRK DDSWRTFDEI MQLARTKDVD MVLLGGDLFH DNKPSRKAMY QVMRSLRKNC LGMKPCELEF LSDPAEVFEG AFPHVNYYDP DINVSIPVFS IHGNHDDPSG DGHLCSLDLL QVAGLVNYFG RVPEADNIHV KPILLQKGKT ...String: MPQTAGPDTI RILVSTDNHV GYEERDPIRK DDSWRTFDEI MQLARTKDVD MVLLGGDLFH DNKPSRKAMY QVMRSLRKNC LGMKPCELEF LSDPAEVFEG AFPHVNYYDP DINVSIPVFS IHGNHDDPSG DGHLCSLDLL QVAGLVNYFG RVPEADNIHV KPILLQKGKT KLALYGMSNV RDERIHRTFR DNKVRFYRPS QQTGDWFNLL TLHQNHYAHT PTGYLSENML PDFLDLVIWG HEHECLIDPK KNPETGFHVM QPGSSIATSL VPGEAVPKHI AILSITGKSF EVEKIPLRTV RPFVIREITL ATDKRFKGLE KKQDNRQEVT KRLMQIVEEM IAEANEMWRS LHEDSQDDED EEQPLPLIRL KVEYSSPEGT KFEVENPQRF SNRFAGKVAN QNDVVHFYRK KTGTTRKPKE GKRELPEGIA EALEDSDSIS VDALVQEFFA QQSLKILPQA PFGDAVNQFV SKDDKHAVEM FVMDSLSSQV RGLLQLDDDK INEGLDSHIE DFRKVMEKNF LSGQQKQAQR RRRFKEKPEG WDSDLNGHWT LQPEAIEELS SSPEPAKEGG RVRPASRITV GDEDNLFEEE EFVQKTTAKR APTTRATRKT AAATRATTAT KASAPAKKSI AAPRGRKRAN PFQDSAEEEE DVIMDDDDDY KPAPPVKAPP PKPARETQTR GAPKTRQTTL NFSQAERPTR TTQKAIEISD DEISEDDAFE SMPARKSKRY UniProtKB: Double-strand break repair protein MRE11 |
-Macromolecule #2: DH domain-containing protein
| Macromolecule | Name: DH domain-containing protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSKIEKLSIL GVRSFGPHHP ETIAFNTPLT LIVGYNGSGK TTVIECLKYA TTGELPPNST RNGAFIHDPD LVGEKEVRAQ VKLSFRSTIG ESYVVTRNIQ LLVQRNNKRT QKTLEGSLLL RNNGERTVIS TRVAELDKLV SEKLGVPPAI LDAVIFCHQD DSLWPMSEPA ...String: MSKIEKLSIL GVRSFGPHHP ETIAFNTPLT LIVGYNGSGK TTVIECLKYA TTGELPPNST RNGAFIHDPD LVGEKEVRAQ VKLSFRSTIG ESYVVTRNIQ LLVQRNNKRT QKTLEGSLLL RNNGERTVIS TRVAELDKLV SEKLGVPPAI LDAVIFCHQD DSLWPMSEPA ALKKRFDEIF EAQKYTKVIE NIRLLKKKKG DELKILKERE VQDKANKERA EKVDRLMAQL TREILEAREK CNELSKQMEE ESAKIKDKYE QANSFLKIMN DLQTKTEKLE YKKDAIVELR SRIEELPDPD EVLRNTLDEY EQTINRIVAD RDHKAAQFHD LQAELKSARD QHTAKAAEQG KHQSDKEKYE RQLVARERMI REAAERHEIR GYNGDLDDRR IAIFNERIQK ILNDKRRELE RLQRENQEEL DRKTAVIAER ESRKQSVIRD RKAAKDRIIS LGKDMASIQG ELSSIDIDEG TEEMLRAEMK ELQARIEAAK ADEQNANLDA QIKEVNEEIW KLESLSAKLA RELVECTRLA SERAQLDLRR KQLAERKREL EIMTNTWNEQ FSTLLGEGWR PETLERDFSD VLKQQQLLVG EHRKKKDATQ QELKQAEYQL SNARNLHNKL TNEMEACMRA VQTAMKEARD LDSAPPVDEY ITMLETDEKE LAEVETALKL YDELKKHYST IKDRALRFNK CYICDRDFTN QEAAKTRLLE KVAKRLGDEE KKELLEDQAA FMKSLDILRA VRVKYDTYQR LSSELPQLSR EIDSETNRRE DLVRRLEDQD LAFREADNKL QEMETLNKHV MKITQLLKDI SDAEKQVERS QQLSNIETRS ADEINEEQTT CAEQTRAAQA KLTKLTAEKQ RLKDLVRQLE VERLQLENKI SSAVQQLERK KRLQESIARH KEDQNQARNA VQEADEELER LEPEIAGARA ALDEARQACR AKEQKVAAER DAIAQTVSEL NMINSEIQEY LDRGGPSSLA ANQRAIANLE TQMANLEGEM RELTVQINKL NKEIDNSDAK KRNIADNLTY RKNLREKDAL EREIAELEAR NAQEDYDRLI KEAHYLEAHR SKLNADRERL MGMMSTKDEE FRRLNEEYEL DLKDAKAKYK ETHIKVETTK AAIEDLGRGM AAVDHAIMQY HSKMMEQINR TIAELWQSTY QGTDIDTIQI RSDVESTTSS DSGTRRNYNY RVSMVKGDTE MDMRGRCSAG QKVLASIIIR LALAESFCAN CGLIALDEPT TNLDSDNIRS LAESLHGIIK ARQAQGNLQL IVITHDEEFL KYMQCSDFCD DFYRVKRDEK QNSVIVRESI TRITE |
-Macromolecule #3: FHA domain-containing protein
| Macromolecule | Name: FHA domain-containing protein / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MWILESELFD GKRLWLRPGK TYLFGRTVAE AGQLTISDKT VSRKHLTIHI DNVPEGGGRN LRSRSNVIVE DLESKKGTLV NGVQIRGQKT TLTEDVNEIK LGLCPKTLKI RWHPIVLSFS FTSKELRADP WTNLRDSLEQ LDIKYSAEYE PTTTHVVSKK RNTSKGLQAL ...String: MWILESELFD GKRLWLRPGK TYLFGRTVAE AGQLTISDKT VSRKHLTIHI DNVPEGGGRN LRSRSNVIVE DLESKKGTLV NGVQIRGQKT TLTEDVNEIK LGLCPKTLKI RWHPIVLSFS FTSKELRADP WTNLRDSLEQ LDIKYSAEYE PTTTHVVSKK RNTSKGLQAL INGRYIVTDS FINAIVQATE IPEGEEGASS ALEQDFEANW PNPLDHLPPR GEEPGNHTTE TYAPDARRQE VFDGYTFIFY EKKQYDNLFP AISAGKGKAL LKEVVPNRTR VDEFVRYVKS VAGEKGLGSF EDGSEGKGVV VVRYTPKGED SAWYAEFFTK FAQQLDHRPI DQKEFLEAIL ACDASMLRRP LEAMSQPVSV SASVEPQSSE KVRPAVEDRK EVEQSAPKQL QPSAEVPATE ESAPAPHRRE RRTGRSRFKG FDFDDDDIII ETPQAQSSTQ VPALPQVPSA SQDSLFVSQR EPSLAPSEPM LEEEAPCNTR TTRQTHRKRV LSPLPEHDNS ALLDEIAPIT AAVKRRRIEA GQDPVPPLPE PEPEREDEDV EMVEESPPRK GKKGAATTAK GKGKKIKQED EENVLELARR RREEAEAAAA AERQRLAQLG DDDIDYAAIR RLHIIEEIEV RQPEPHGPNR TREQDIADGR WDPRWNGRKN FKRFRRQGET GVRMPVQSVI VPLEEVRTKE YGIGDDYWLE DEEGRVPRRP KETQTQERST IGSVRDGSGF AAAAASGKGK EKDKENEKEV GRPGSSAAAA KQRSKPAPRR TVLTLDSSDE DEDEPSPHAP GIDTISDSEP EVVSSFPSVI PASEPSRSRA AKAAERANAL RSSAHSSQSQ TQQHRESQLS TGSSKIQLTL APGSSSLSFS RSGTAAGRNE NGKRPFGSFV SGESTASGRG MSVESGSVRG ESASKRQKQG SSGGGSFLAT RRKDDGSEEE SEDDELKFRF GRRR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.27 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)