+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13206 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tetrameric building block of the human GID complex. | |||||||||
 Map data Map data | Postprocessed map of the four building blocks of the tetrameric hGID complex, consisting of WDR26, RanBP9, TWA1, and MAEA or RMND5A. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Mohamed WI / Park SL / Rabl J / Leitner A / Boehringer D / Peter M | |||||||||
| Funding support | European Union,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2021 Journal: EMBO Rep / Year: 2021Title: The human GID complex engages two independent modules for substrate recruitment. Authors: Weaam I Mohamed / Sophia L Park / Julius Rabl / Alexander Leitner / Daniel Boehringer / Matthias Peter /  Abstract: The human GID (hGID) complex is a conserved E3 ubiquitin ligase regulating diverse biological processes, including glucose metabolism and cell cycle progression. However, the biochemical function and ...The human GID (hGID) complex is a conserved E3 ubiquitin ligase regulating diverse biological processes, including glucose metabolism and cell cycle progression. However, the biochemical function and substrate recognition of the multi-subunit complex remain poorly understood. Using biochemical assays, cross-linking mass spectrometry, and cryo-electron microscopy, we show that hGID engages two distinct modules for substrate recruitment, dependent on either WDR26 or GID4. WDR26 and RanBP9 cooperate to ubiquitinate HBP1 in vitro, while GID4 is dispensable for this reaction. In contrast, GID4 functions as an adaptor for the substrate ZMYND19, which surprisingly lacks a Pro/N-end degron. GID4 substrate binding and ligase activity is regulated by ARMC8α, while the shorter ARMC8β isoform assembles into a stable hGID complex that is unable to recruit GID4. Cryo-EM reconstructions of these hGID complexes reveal the localization of WDR26 within a ring-like, tetrameric architecture and suggest that GID4 and WDR26/Gid7 utilize different, non-overlapping binding sites. Together, these data advance our mechanistic understanding of how the hGID complex recruits cognate substrates and provides insights into the regulation of its E3 ligase activity. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: The human GID complex engages two independent modules for substrate recruitment Authors: Mohamed WI / Park SL / Rabl J / Leitner A / Boehringer D / Peter M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13206.map.gz emd_13206.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13206-v30.xml emd-13206-v30.xml emd-13206.xml emd-13206.xml | 27.6 KB 27.6 KB | Display Display |  EMDB header EMDB header |
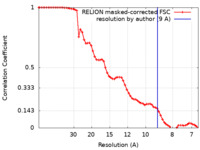
| FSC (resolution estimation) |  emd_13206_fsc.xml emd_13206_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_13206.png emd_13206.png | 47.7 KB | ||
| Masks |  emd_13206_msk_1.map emd_13206_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Others |  emd_13206_additional_1.map.gz emd_13206_additional_1.map.gz emd_13206_half_map_1.map.gz emd_13206_half_map_1.map.gz emd_13206_half_map_2.map.gz emd_13206_half_map_2.map.gz | 17.1 MB 17.1 MB 17.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13206 http://ftp.pdbj.org/pub/emdb/structures/EMD-13206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13206 | HTTPS FTP |
-Validation report
| Summary document |  emd_13206_validation.pdf.gz emd_13206_validation.pdf.gz | 422.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13206_full_validation.pdf.gz emd_13206_full_validation.pdf.gz | 422 KB | Display | |
| Data in XML |  emd_13206_validation.xml.gz emd_13206_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_13206_validation.cif.gz emd_13206_validation.cif.gz | 16.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13206 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13206 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13206 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13206 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13206.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13206.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map of the four building blocks of the tetrameric hGID complex, consisting of WDR26, RanBP9, TWA1, and MAEA or RMND5A. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13206_msk_1.map emd_13206_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_13206_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13206_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13206_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : hGID
| Entire | Name: hGID |
|---|---|
| Components |
|
-Supramolecule #1: hGID
| Supramolecule | Name: hGID / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Complex of subunits WDR26, RanBP9, TWA1, MAEA, and RMND5A. The map contains one subunit of WDR26, RanBP9, TWA1, and one subunit of MAEA or RMND5A. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Macromolecule #1: RanBP9
| Macromolecule | Name: RanBP9 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MDWSHPQFEK SAVDENLYFQ GGGRMSGQPP PPPPQQQQQQ QQLSPPPPAA LAPVSGVVLP APPAVSAGSS PAGSPGGGAG GEGLGAAAAA LLLHPPPPPP PATAAPPPPP PPPPPPASAA APASGPPAPP GLAAGPGPAG GAPTPALVAG SSAAAPFPHG DSALNEQEKE ...String: MDWSHPQFEK SAVDENLYFQ GGGRMSGQPP PPPPQQQQQQ QQLSPPPPAA LAPVSGVVLP APPAVSAGSS PAGSPGGGAG GEGLGAAAAA LLLHPPPPPP PATAAPPPPP PPPPPPASAA APASGPPAPP GLAAGPGPAG GAPTPALVAG SSAAAPFPHG DSALNEQEKE LQRRLKRLYP AVDEQETPLP RSWSPKDKFS YIGLSQNNLR VHYKGHGKTP KDAASVRATH PIPAACGIYY FEVKIVSKGR DGYMGIGLSA QGVNMNRLPG WDKHSYGYHG DDGHSFCSSG TGQPYGPTFT TGDVIGCCVN LINNTCFYTK NGHSLGIAFT DLPPNLYPTV GLQTPGEVVD ANFGQHPFVF DIEDYMREWR TKIQAQIDRF PIGDREGEWQ TMIQKMVSSY LVHHGYCATA EAFARSTDQT VLEELASIKN RQRIQKLVLA GRMGEAIETT QQLYPSLLER NPNLLFTLKV RQFIEMVNGT DSEVRCLGGR SPKSQDSYPV SPRPFSSPSM SPSHGMNIHN LASGKGSTAH FSGFESCSNG VISNKAHQSY CHSNKHQSSN LNVPELNSIN MSRSQQVNNF TSNDVDMETD HYSNGVGETS SNGFLNGSSK HDHEMEDCDT EMEVDSSQLR RQLCGGSQAA IERMIHFGRE LQAMSEQLRR DCGKNTANKK MLKDAFSLLA YSDPWNSPVG NQLDPIQREP VCSALNSAIL ETHNLPKQPP LALAMGQATQ CLGLMARSGI GSCAFATVED YLH |
-Macromolecule #2: WDR26
| Macromolecule | Name: WDR26 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SAVDENLYFQ GGGRSQSDED VIRLIGQHLN GLGLNQTVDL LMQESGCRLE HPSATKFRNH VMEGDWDKAE NDLNELKPLV HSPHAIVVRG ALEISQTLLG IIVRMKFLLL QQKYLEYLED GKVLEALQVL RCELTPLKYN TERIHVLSGY LMCSHAEDLR ...String: MGSSHHHHHH SAVDENLYFQ GGGRSQSDED VIRLIGQHLN GLGLNQTVDL LMQESGCRLE HPSATKFRNH VMEGDWDKAE NDLNELKPLV HSPHAIVVRG ALEISQTLLG IIVRMKFLLL QQKYLEYLED GKVLEALQVL RCELTPLKYN TERIHVLSGY LMCSHAEDLR AKAEWEGKGT ASRSKLLDKL QTYLPPSVML PPRRLQTLLR QAVELQRDRC LYHNTKLDNN LDSVSLLIDH VCSRRQFPCY TQQILTEHCN EVWFCKFSND GTKLATGSKD TTVIIWQVDP DTHLLKLLKT LEGHAYGVSY IAWSPDDNYL VACGPDDCSE LWLWNVQTGE LRTKMSQSHE DSLTSVAWNP DGKRFVTGGQ RGQFYQCDLD GNLLDSWEGV RVQCLWCLSD GKTVLASDTH QRIRGYNFED LTDRNIVQED HPIMSFTISK NGRLALLNVA TQGVHLWDLQ DRVLVRKYQG VTQGFYTIHS CFGGHNEDFI ASGSEDHKVY IWHKRSELPI AELTGHTRTV NCVSWNPQIP SMMASASDDG TVRIWGPAPF IDHQNIEEEC SSMDS |
-Macromolecule #3: Twa1
| Macromolecule | Name: Twa1 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SAVDENLYFQ GGGRMSYAEK PDEITKDEWM EKLNNLHVQR ADMNRLIMNY LVTEGFKEAA EKFRMESGIE PSVDLETLDE RIKIREMILK GQIQEAIALI NSLHPELLDT NRYLYFHLQQ QHLIELIRQR ETEAALEFAQ TQLAEQGEES RECLTEMERT ...String: MGSSHHHHHH SAVDENLYFQ GGGRMSYAEK PDEITKDEWM EKLNNLHVQR ADMNRLIMNY LVTEGFKEAA EKFRMESGIE PSVDLETLDE RIKIREMILK GQIQEAIALI NSLHPELLDT NRYLYFHLQQ QHLIELIRQR ETEAALEFAQ TQLAEQGEES RECLTEMERT LALLAFDSPE ESPFGDLLHT MQRQKVWSEV NQAVLDYENR ESTPKLAKLL KLLLWAQNEL DQKKVKYPKM TDLSKGVIEE PK |
-Macromolecule #4: MAEA
| Macromolecule | Name: MAEA / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKS AVDENLYFQG GGRMAVQESA AQLSMTLKVQ EYPTLKVPYE TLNKRFRAAQ KNIDRETSHV TMVVAELEKT LSGCPAVDSV VSLLDGVVEK LSVLKRKAVE SIQAEDESAK LCKRRIEHLK EHSSDQPAAA SVWKRKRMDR MMVEHLLRCG YYNTAVKLAR ...String: MDYKDDDDKS AVDENLYFQG GGRMAVQESA AQLSMTLKVQ EYPTLKVPYE TLNKRFRAAQ KNIDRETSHV TMVVAELEKT LSGCPAVDSV VSLLDGVVEK LSVLKRKAVE SIQAEDESAK LCKRRIEHLK EHSSDQPAAA SVWKRKRMDR MMVEHLLRCG YYNTAVKLAR QSGIEDLVNI EMFLTAKEVE ESLERRETAT CLAWCHDNKS RLRKMKSCLE FSLRIQEFIE LIRQNKRLDA VRHARKHFSQ AEGSQLDEVR QAMGMLAFPP DTHISPYKDL LDPARWRMLI QQFRYDNYRL HQLGNNSVFT LTLQAGLSAI KTPQCYKEDG SSKSPDCPVC SRSLNKLAQP LPMAHCANSR LVCKISGDVM NENNPPMMLP NGYVYGYNSL LSIRQDDKVV CPRTKEVFHF SQAEKVYIM |
-Macromolecule #5: RMND5A
| Macromolecule | Name: RMND5A / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SAVDENLYFQ GGGRMDQCVT VERELEKVLH KFSGYGQLCE RGLEELIDYT GGLKHEILQS HGQDAELSGT LSLVLTQCCK RIKDTVQKLA SDHKDIHSSV SRVGKAIDKN FDSDISSVGI DGCWQADSQR LLNEVMVEHF FRQGMLDVAE ELCQESGLSV ...String: MGSSHHHHHH SAVDENLYFQ GGGRMDQCVT VERELEKVLH KFSGYGQLCE RGLEELIDYT GGLKHEILQS HGQDAELSGT LSLVLTQCCK RIKDTVQKLA SDHKDIHSSV SRVGKAIDKN FDSDISSVGI DGCWQADSQR LLNEVMVEHF FRQGMLDVAE ELCQESGLSV DPSQKEPFVE LNRILEALKV RVLRPALEWA VSNREMLIAQ NSSLEFKLHR LYFISLLMGG TTNQREALQY AKNFQPFALN HQKDIQVLMG SLVYLRQGIE NSPYVHLLDA NQWADICDIF TRDACALLGL SVESPLSVSF SAGCVALPAL INIKAVIEQR QCTGVWNQKD ELPIEVDLGK KCWYHSIFAC PILRQQTTDN NPPMKLVCGH IISRDALNKM FNGSKLKCPY CPMEQSPGDA KQIFF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 50 mM HEPES pH 7.4, 200 mM NaCl, 1mM TCEP and 0.01% NP40 | |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 1.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: 15mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | The complex was purified with gel filtration and subsequently crosslinked using the GraFix method. |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 26017 / Average exposure time: 1.5 sec. / Average electron dose: 78.0 e/Å2 / Details: magnification: 105000x, 0.84A/pix |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 19599 / Average exposure time: 8.5 sec. / Average electron dose: 78.0 e/Å2 / Details: magnification: 165000, 0.84A/pix |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)