+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12799 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SRP-SR at the distal site conformation | |||||||||
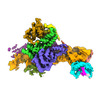
 Map data Map data | SRP-SR complex from 3D focused refinement on ribosome signal subtracted images | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SRP SRP receptor / co-translational protein targeting / endoplasmic reticulum / signal peptide / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationSRP-dependent cotranslational protein targeting to membrane / signal recognition particle receptor complex / SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition / endoplasmic reticulum signal peptide binding / signal recognition particle, endoplasmic reticulum targeting / granulocyte differentiation / signal recognition particle binding / protein targeting to ER / signal-recognition-particle GTPase / exocrine pancreas development ...SRP-dependent cotranslational protein targeting to membrane / signal recognition particle receptor complex / SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition / endoplasmic reticulum signal peptide binding / signal recognition particle, endoplasmic reticulum targeting / granulocyte differentiation / signal recognition particle binding / protein targeting to ER / signal-recognition-particle GTPase / exocrine pancreas development / SRP-dependent cotranslational protein targeting to membrane, translocation / 7S RNA binding / SRP-dependent cotranslational protein targeting to membrane / cytoplasmic microtubule / aminopeptidase activity / membrane => GO:0016020 / vesicle-mediated transport / neutrophil chemotaxis / intracellular protein transport / GDP binding / nuclear speck / serine-type endopeptidase activity / GTPase activity / endoplasmic reticulum membrane / GTP binding / nucleolus / Golgi apparatus / endoplasmic reticulum / ATP hydrolysis activity / nucleoplasm / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Jomaa A / Ban N | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle. Authors: Ahmad Jomaa / Simon Eitzinger / Zikun Zhu / Sowmya Chandrasekar / Kan Kobayashi / Shu-Ou Shan / Nenad Ban /   Abstract: Co-translational protein targeting to membranes by the signal recognition particle (SRP) is a universally conserved pathway from bacteria to humans. In mammals, SRP and its receptor (SR) have many ...Co-translational protein targeting to membranes by the signal recognition particle (SRP) is a universally conserved pathway from bacteria to humans. In mammals, SRP and its receptor (SR) have many additional RNA features and protein components compared to the bacterial system, which were recently shown to play regulatory roles. Due to its complexity, the mammalian SRP targeting process is mechanistically not well understood. In particular, it is not clear how SRP recognizes translating ribosomes with exposed signal sequences and how the GTPase activity of SRP and SR is regulated. Here, we present electron cryo-microscopy structures of SRP and SRP·SR in complex with the translating ribosome. The structures reveal the specific molecular interactions between SRP and the emerging signal sequence and the elements that regulate GTPase activity of SRP·SR. Our results suggest the molecular mechanism of how eukaryote-specific elements regulate the early and late stages of SRP-dependent protein targeting. #1:  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle Authors: Jomaa A / Eitzinger S / Zhu Z / Chandrasekar S / Kobayashi K / Shan S / Ban N | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12799.map.gz emd_12799.map.gz | 320.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12799-v30.xml emd-12799-v30.xml emd-12799.xml emd-12799.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12799.png emd_12799.png | 87 KB | ||
| Filedesc metadata |  emd-12799.cif.gz emd-12799.cif.gz | 8.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12799 http://ftp.pdbj.org/pub/emdb/structures/EMD-12799 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12799 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12799 | HTTPS FTP |
-Validation report
| Summary document |  emd_12799_validation.pdf.gz emd_12799_validation.pdf.gz | 546.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12799_full_validation.pdf.gz emd_12799_full_validation.pdf.gz | 546.4 KB | Display | |
| Data in XML |  emd_12799_validation.xml.gz emd_12799_validation.xml.gz | 7.3 KB | Display | |
| Data in CIF |  emd_12799_validation.cif.gz emd_12799_validation.cif.gz | 8.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12799 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12799 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12799 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12799 | HTTPS FTP |
-Related structure data
| Related structure data |  7obqMC  7obrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12799.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12799.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SRP-SR complex from 3D focused refinement on ribosome signal subtracted images | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : SRP-SR complex
+Supramolecule #1: SRP-SR complex
+Supramolecule #2: SRP-SR complex
+Supramolecule #3: EM14S01-3B_G0054400.mRNA.1.CDS.1
+Supramolecule #4: Signal recognition particle receptor subunit
+Macromolecule #1: SRP RNA
+Macromolecule #2: Signal recognition particle 19
+Macromolecule #3: EM14S01-3B_G0054400.mRNA.1.CDS.1
+Macromolecule #4: Signal recognition particle subunit SRP68,Signal recognition part...
+Macromolecule #5: Signal recognition particle receptor subunit beta
+Macromolecule #6: Signal recognition particle 54 kDa protein
+Macromolecule #7: SRP receptor subunit alpha
+Macromolecule #8: Signal recognition particle subunit SRP72
+Macromolecule #9: MAGNESIUM ION
+Macromolecule #10: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #11: PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: From initial reconstruction |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 155989 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)

























