[English] 日本語
 Yorodumi
Yorodumi- EMDB-12307: P1b-state of wild type human mitochondrial LONP1 protease with bo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12307 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | P1b-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | human mitochondrial AAA+ protease / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationoxidation-dependent protein catabolic process / response to aluminum ion / PH domain binding / endopeptidase La / mitochondrial protein catabolic process / G-quadruplex DNA binding / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / mitochondrial nucleoid / insulin receptor substrate binding ...oxidation-dependent protein catabolic process / response to aluminum ion / PH domain binding / endopeptidase La / mitochondrial protein catabolic process / G-quadruplex DNA binding / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / mitochondrial nucleoid / insulin receptor substrate binding / Mitochondrial unfolded protein response (UPRmt) / chaperone-mediated protein complex assembly / response to hormone / DNA polymerase binding / negative regulation of insulin receptor signaling pathway / Mitochondrial protein degradation / : / mitochondrion organization / ADP binding / single-stranded DNA binding / cellular response to oxidative stress / sequence-specific DNA binding / response to hypoxia / single-stranded RNA binding / mitochondrial matrix / serine-type endopeptidase activity / ATP hydrolysis activity / mitochondrion / nucleoplasm / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
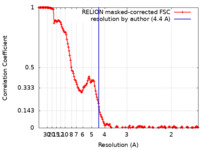
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Mohammed I / Schmitz KA | |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Catalytic cycling of human mitochondrial Lon protease. Authors: Inayathulla Mohammed / Kai A Schmitz / Niko Schenck / Dimitrios Balasopoulos / Annika Topitsch / Timm Maier / Jan Pieter Abrahams /  Abstract: The mitochondrial Lon protease (LonP1) regulates mitochondrial health by removing redundant proteins from the mitochondrial matrix. We determined LonP1 in eight nucleotide-dependent conformational ...The mitochondrial Lon protease (LonP1) regulates mitochondrial health by removing redundant proteins from the mitochondrial matrix. We determined LonP1 in eight nucleotide-dependent conformational states by cryoelectron microscopy (cryo-EM). The flexible assembly of N-terminal domains had 3-fold symmetry, and its orientation depended on the conformational state. We show that a conserved structural motif around T803 with a high similarity to the trypsin catalytic triad is essential for proteolysis. We show that LonP1 is not regulated by redox potential, despite the presence of two conserved cysteines at disulfide-bonding distance in its unfoldase core. Our data indicate how sequential ATP hydrolysis controls substrate protein translocation in a 6-fold binding change mechanism. Substrate protein translocation, rather than ATP hydrolysis, is a rate-limiting step, suggesting that LonP1 is a Brownian ratchet with ATP hydrolysis preventing translocation reversal. 3-fold rocking motions of the flexible N-domain assembly may assist thermal unfolding of the substrate protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12307.map.gz emd_12307.map.gz | 323.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12307-v30.xml emd-12307-v30.xml emd-12307.xml emd-12307.xml | 27.4 KB 27.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12307_fsc.xml emd_12307_fsc.xml | 16 KB | Display |  FSC data file FSC data file |
| Images |  emd_12307.png emd_12307.png | 64.7 KB | ||
| Masks |  emd_12307_msk_1.map emd_12307_msk_1.map | 343 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12307.cif.gz emd-12307.cif.gz | 7.7 KB | ||
| Others |  emd_12307_half_map_1.map.gz emd_12307_half_map_1.map.gz emd_12307_half_map_2.map.gz emd_12307_half_map_2.map.gz | 318.2 MB 318.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12307 http://ftp.pdbj.org/pub/emdb/structures/EMD-12307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12307 | HTTPS FTP |
-Related structure data
| Related structure data |  7ng4MC  7nfyC  7ng5C  7ngcC  7ngfC  7nglC  7ngpC  7ngqC  7oxoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12307.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12307.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
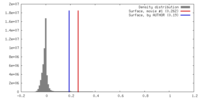
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12307_msk_1.map emd_12307_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
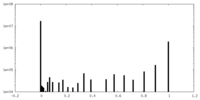
| Projections & Slices |
| ||||||||||||
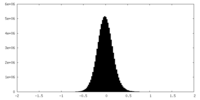
| Density Histograms |
-Half map: #2
| File | emd_12307_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
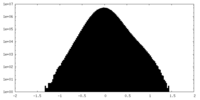
| Density Histograms |
-Half map: #1
| File | emd_12307_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : P1b-state of hexameric LONP1 complex with ATP/ADP and bound endog...
| Entire | Name: P1b-state of hexameric LONP1 complex with ATP/ADP and bound endogenous substrate protein |
|---|---|
| Components |
|
-Supramolecule #1: P1b-state of hexameric LONP1 complex with ATP/ADP and bound endog...
| Supramolecule | Name: P1b-state of hexameric LONP1 complex with ATP/ADP and bound endogenous substrate protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: Lon protease homolog, mitochondrial
| Macromolecule | Name: Lon protease homolog, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 96.288758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHGSMT IPDVFPHLPL IAITRNPVFP RFIKIIEVKN KKLVELLRRK VRLAQPYVGV FLKRDDSNES DVVESLDEIY HTGTFAQIH EMQDLGDKLR MIVMGHRRVH ISRQLEVEPE EPEAENKHKP RRKSKRGKKE AEDELSARHP AELAMEPTPE L PAEVLMVE ...String: HHHHHHGSMT IPDVFPHLPL IAITRNPVFP RFIKIIEVKN KKLVELLRRK VRLAQPYVGV FLKRDDSNES DVVESLDEIY HTGTFAQIH EMQDLGDKLR MIVMGHRRVH ISRQLEVEPE EPEAENKHKP RRKSKRGKKE AEDELSARHP AELAMEPTPE L PAEVLMVE VENVVHEDFQ VTEEVKALTA EIVKTIRDII ALNPLYRESV LQMMQAGQRV VDNPIYLSDM GAALTGAESH EL QDVLEET NIPKRLYKAL SLLKKEFELS KLQQRLGREV EEKIKQTHRK YLLQEQLKII KKELGLEKDD KDAIEEKFRE RLK ELVVPK HVMDVVDEEL SKLGLLDNHS SEFNVTRNYL DWLTSIPWGK YSNENLDLAR AQAVLEEDHY GMEDVKKRIL EFIA VSQLR GSTQGKILCF YGPPGVGKTS IARSIARALN REYFRFSVGG MTDVAEIKGH RRTYVGAMPG KIIQCLKKTK TENPL ILID EVDKIGRGYQ GDPSSALLEL LDPEQNANFL DHYLDVPVDL SKVLFICTAN VTDTIPEPLR DRMEMINVSG YVAQEK LAI AERYLVPQAR ALCGLDESKA KLSSDVLTLL IKQYCRESGV RNLQKQVEKV LRKSAYKIVS GEAESVEVTP ENLQDFV GK PVFTVERMYD VTPPGVVMGL AWTAMGGSTL FVETSLRRPQ DKDAKGDKDG SLEVTGQLGE VMKESARIAY TFARAFLM Q HAPANDYLVT SHIHLHVPEG ATPKDGPSAG CTIVTALLSL AMGRPVRQNL AMTGEVSLTG KILPVGGIKE KTIAAKRAG VTCIVLPAEN KKDFYDLAAF ITEGLEVHFV EHYREIFDIA FPDEQAEALA VER UniProtKB: Lon protease homolog, mitochondrial |
-Macromolecule #2: substrate protein, chain G
| Macromolecule | Name: substrate protein, chain G / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.698783 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.45 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7ng4: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)