+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11783 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | OpuA (E190Q) occluded | ||||||||||||
 Map data Map data | OpuA (E190Q) %u2013 Glycine betaine, ATP at 3.3A resolution | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | osmoregulation / ABC-transporter / glycine betaine uptake system / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type quaternary amine transporter / amine transmembrane transporter activity / ABC-type quaternary ammonium compound transporting activity / carnitine transmembrane transporter activity / glycine betaine transport / choline transport / peptide transport / amino acid transport / ATP-binding cassette (ABC) transporter complex / protein transport ...ABC-type quaternary amine transporter / amine transmembrane transporter activity / ABC-type quaternary ammonium compound transporting activity / carnitine transmembrane transporter activity / glycine betaine transport / choline transport / peptide transport / amino acid transport / ATP-binding cassette (ABC) transporter complex / protein transport / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Lactococcus lactis subsp. lactis (lactic acid bacteria) Lactococcus lactis subsp. lactis (lactic acid bacteria) | ||||||||||||
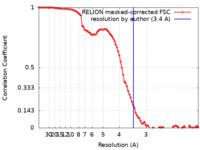
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Sikkema HR / Rheinberger J | ||||||||||||
| Funding support |  Netherlands, 3 items Netherlands, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2020 Journal: Sci Adv / Year: 2020Title: Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA. Authors: Hendrik R Sikkema / Marco van den Noort / Jan Rheinberger / Marijn de Boer / Sabrina T Krepel / Gea K Schuurman-Wolters / Cristina Paulino / Bert Poolman /  Abstract: (Micro)organisms are exposed to fluctuating environmental conditions, and adaptation to stress is essential for survival. Increased osmolality (hypertonicity) causes outflow of water and loss of ...(Micro)organisms are exposed to fluctuating environmental conditions, and adaptation to stress is essential for survival. Increased osmolality (hypertonicity) causes outflow of water and loss of turgor and is dangerous if the cell is not capable of rapidly restoring its volume. The osmoregulatory adenosine triphosphate-binding cassette transporter OpuA restores the cell volume by accumulating large amounts of compatible solute. OpuA is gated by ionic strength and inhibited by the second messenger cyclic-di-AMP, a molecule recently shown to affect many cellular processes. Despite the master regulatory role of cyclic-di-AMP, structural and functional insights into how the second messenger regulates (transport) proteins on the molecular level are lacking. Here, we present high-resolution cryo-electron microscopy structures of OpuA and in vitro activity assays that show how the osmoregulator OpuA is activated by high ionic strength and how cyclic-di-AMP acts as a backstop to prevent unbridled uptake of compatible solutes. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11783.map.gz emd_11783.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11783-v30.xml emd-11783-v30.xml emd-11783.xml emd-11783.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11783_fsc.xml emd_11783_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11783.png emd_11783.png | 161.5 KB | ||
| Masks |  emd_11783_msk_1.map emd_11783_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11783.cif.gz emd-11783.cif.gz | 6.7 KB | ||
| Others |  emd_11783_half_map_1.map.gz emd_11783_half_map_1.map.gz emd_11783_half_map_2.map.gz emd_11783_half_map_2.map.gz | 49.8 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11783 http://ftp.pdbj.org/pub/emdb/structures/EMD-11783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11783 | HTTPS FTP |
-Validation report
| Summary document |  emd_11783_validation.pdf.gz emd_11783_validation.pdf.gz | 880.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11783_full_validation.pdf.gz emd_11783_full_validation.pdf.gz | 880 KB | Display | |
| Data in XML |  emd_11783_validation.xml.gz emd_11783_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  emd_11783_validation.cif.gz emd_11783_validation.cif.gz | 21.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11783 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11783 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11783 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11783 | HTTPS FTP |
-Related structure data
| Related structure data |  7ahdMC  7ahcC  7aheC  7ahhC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11783.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11783.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | OpuA (E190Q) %u2013 Glycine betaine, ATP at 3.3A resolution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.012 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
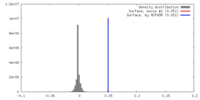
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11783_msk_1.map emd_11783_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
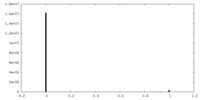
| Density Histograms |
-Half map: #2
| File | emd_11783_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
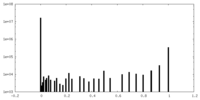
| Density Histograms |
-Half map: #1
| File | emd_11783_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
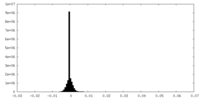
| Density Histograms |
- Sample components
Sample components
-Entire : OpuA (E190Q) occluded
| Entire | Name: OpuA (E190Q) occluded |
|---|---|
| Components |
|
-Supramolecule #1: OpuA (E190Q) occluded
| Supramolecule | Name: OpuA (E190Q) occluded / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis subsp. lactis (lactic acid bacteria) Lactococcus lactis subsp. lactis (lactic acid bacteria) |
-Macromolecule #1: ABC-type proline/glycine betaine transport system ATPase component
| Macromolecule | Name: ABC-type proline/glycine betaine transport system ATPase component type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis subsp. lactis (lactic acid bacteria) Lactococcus lactis subsp. lactis (lactic acid bacteria) |
| Molecular weight | Theoretical: 45.782699 KDa |
| Recombinant expression | Organism:  Lactococcus lactis subsp. lactis (lactic acid bacteria) Lactococcus lactis subsp. lactis (lactic acid bacteria) |
| Sequence | String: MAVKIKIEHL TKIFGKRIKT ALTMVEKGEP KNEILKKTGA TVGVYDTNFE INEGEIFVIM GLSGSGKSTL LRLLNRLIEP TSGKIFIDN QDVATLNKED LLQVRRKTMS MVFQNFGLFP HRTILENTEY GLEVQNVPKE ERRKRAEKAL DNANLLDFKD Q YPKQLSGG ...String: MAVKIKIEHL TKIFGKRIKT ALTMVEKGEP KNEILKKTGA TVGVYDTNFE INEGEIFVIM GLSGSGKSTL LRLLNRLIEP TSGKIFIDN QDVATLNKED LLQVRRKTMS MVFQNFGLFP HRTILENTEY GLEVQNVPKE ERRKRAEKAL DNANLLDFKD Q YPKQLSGG MQQRVGLARA LANDPEILLM DQAFSALDPL IRREMQDELL ELQAKFQKTI IFVSHDLNEA LRIGDRIAIM KD GKIMQIG TGEEILTNPA NDYVKTFVED VDRAKVITAE NIMIPALTTN IDVDGPSVAL KKMKTEEVSS LMAVDKKRQF RGV VTSEQA IAARKNNQPL KDVMTTDVGT VSKEMLVRDI LPIIYDAPTP LAVVDDNGFL KGVLIRGSVL EALADIPDED EVEE IEKEE ENK UniProtKB: Quaternary amine transport ATP-binding protein |
-Macromolecule #2: ABC transporter permease subunit
| Macromolecule | Name: ABC transporter permease subunit / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis subsp. lactis (lactic acid bacteria) Lactococcus lactis subsp. lactis (lactic acid bacteria) |
| Molecular weight | Theoretical: 63.437812 KDa |
| Recombinant expression | Organism:  Lactococcus lactis subsp. lactis (lactic acid bacteria) Lactococcus lactis subsp. lactis (lactic acid bacteria) |
| Sequence | String: MIDLAIGQVP IANWVSSATD WITSTFSSGF DVIQKSGTVL MNGITGALTA VPFWLMIAVV TILAILVSGK KIAFPLFTFI GLSLIANQG LWSDLMSTIT LVLLSSLLSI IIGVPLGIWM AKSDLVAKIV QPILDFMQTM PGFVYLIPAV AFFGIGVVPG V FASVIFAL ...String: MIDLAIGQVP IANWVSSATD WITSTFSSGF DVIQKSGTVL MNGITGALTA VPFWLMIAVV TILAILVSGK KIAFPLFTFI GLSLIANQG LWSDLMSTIT LVLLSSLLSI IIGVPLGIWM AKSDLVAKIV QPILDFMQTM PGFVYLIPAV AFFGIGVVPG V FASVIFAL PPTVRMTNLG IRQVSTELVE AADSFGSTAR QKLFKLEFPL AKGTIMAGVN QTIMLALSMV VIASMIGAPG LG RGVLAAV QSADIGKGFV SGISLVILAI IIDRFTQKLN VSPLEKQGNP TVKKWKRGIA LVSLLALIIG AFSGMSFGKT ASD KKVDLV YMNWDSEVAS INVLTQAMKE HGFDVKTTAL DNAVAWQTVA NGQADGMVSA WLPNTHKTQW QKYGKSVDLL GPNL KGAKV GFVVPSYMNV NSIEDLTNQA NKTITGIEPG AGVMAASEKT LNSYDNLKDW KLVPSSSGAM TVALGEAIKQ HKDIV ITGW SPHWMFNKYD LKYLADPKGT MGTSENINTI VRKGLKKENP EAYKVLDKFN WTTKDMEAVM LDIQNGKTPE EAAKNW IKD HQKEVDKWFK GSIEGRHHHH HH UniProtKB: ABC transporter permease/substrate binding protein |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: TRIMETHYL GLYCINE
| Macromolecule | Name: TRIMETHYL GLYCINE / type: ligand / ID: 4 / Number of copies: 1 / Formula: BET |
|---|---|
| Molecular weight | Theoretical: 118.154 Da |
| Chemical component information |  ChemComp-BET: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Details: at 5mA | |||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 90.0 K / Max: 105.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 2 / Number real images: 2608 / Average exposure time: 9.0 sec. / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 49407 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 49407 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-7ahd: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)