[English] 日本語
 Yorodumi
Yorodumi- EMDB-1151: Electron cryotomography of the E. coli pyruvate and 2-oxoglutarat... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1151 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryotomography of the E. coli pyruvate and 2-oxoglutarate dehydrogenase complexes. | |||||||||
 Map data Map data | Volume of an extracted E. coli pyruvate dehydrogenase multienzyme complex particle from a dual-tilt tomogram. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Pyruvate dehydrogenase E1 component / Dihydrolipoamide acetyltransferase pyruvate dehydrogenase complex / Dihydrolipoamide dehydrogenase Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 55.0 Å | |||||||||
 Authors Authors | Murphy GE / Jensen GJ | |||||||||
 Citation Citation |  Journal: Structure / Year: 2005 Journal: Structure / Year: 2005Title: Electron cryotomography of the E. coli pyruvate and 2-oxoglutarate dehydrogenase complexes. Authors: Gavin E Murphy / Grant J Jensen /  Abstract: The E. coli pyruvate and 2-oxoglutarate dehydrogenases are two closely related, large complexes that exemplify a growing number of multiprotein "machines" whose domains have been studied extensively ...The E. coli pyruvate and 2-oxoglutarate dehydrogenases are two closely related, large complexes that exemplify a growing number of multiprotein "machines" whose domains have been studied extensively and modeled in atomic detail, but whose quaternary structures have remained unclear for lack of an effective imaging technology. Here, electron cryotomography was used to show that the E1 and E3 subunits of these complexes are flexibly tethered approximately 11 nm away from the E2 core. This result demonstrates unambiguously that electron cryotomography can reveal the relative positions of features as small as 80 kDa in individual complexes, elucidating quaternary structure and conformational flexibility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1151.map.gz emd_1151.map.gz | 224.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1151-v30.xml emd-1151-v30.xml emd-1151.xml emd-1151.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  1151.gif 1151.gif | 21.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1151 http://ftp.pdbj.org/pub/emdb/structures/EMD-1151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1151 | HTTPS FTP |
-Validation report
| Summary document |  emd_1151_validation.pdf.gz emd_1151_validation.pdf.gz | 246.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1151_full_validation.pdf.gz emd_1151_full_validation.pdf.gz | 245.7 KB | Display | |
| Data in XML |  emd_1151_validation.xml.gz emd_1151_validation.xml.gz | 4.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1151 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1151 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1151 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1151 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1151.map.gz / Format: CCP4 / Size: 489.3 KB / Type: IMAGE STORED AS SIGNED BYTE Download / File: emd_1151.map.gz / Format: CCP4 / Size: 489.3 KB / Type: IMAGE STORED AS SIGNED BYTE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Volume of an extracted E. coli pyruvate dehydrogenase multienzyme complex particle from a dual-tilt tomogram. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
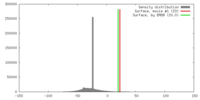
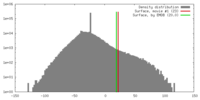
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Unengineered E. coli Pyruvate Dehydrogenase Multienzyme Complex
| Entire | Name: Unengineered E. coli Pyruvate Dehydrogenase Multienzyme Complex |
|---|---|
| Components |
|
-Supramolecule #1000: Unengineered E. coli Pyruvate Dehydrogenase Multienzyme Complex
| Supramolecule | Name: Unengineered E. coli Pyruvate Dehydrogenase Multienzyme Complex type: sample / ID: 1000 Details: The sample was thawed from storage at -80 degrees Celcius before being loaded onto the grid. Oligomeric state: Up to 24 pyruvate dehydrogenase E1p and dihydrolipoamide dehydrogenase E3 dimers together bind to the 24 dihydrolipoamide acetyltransferase E2p octahedral core. Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 5.6 MDa |
-Macromolecule #1: E2p octahedral core
| Macromolecule | Name: E2p octahedral core / type: protein_or_peptide / ID: 1 / Name.synonym: dihydrolipoamide acetyltransferase Details: 24 arranged as cube; See Wagenknecht, T. et al., JSB 109:70-77 (1992). Number of copies: 24 / Oligomeric state: 24mer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 1.6 MDa |
| Recombinant expression | Organism:  |
| Sequence | InterPro: Dihydrolipoamide acetyltransferase pyruvate dehydrogenase complex |
-Macromolecule #2: E1p
| Macromolecule | Name: E1p / type: protein_or_peptide / ID: 2 / Name.synonym: pyruvate dehydrogenase Details: Up to 24 of these; See Wagenknecht, T. et al., JSB 109:70-77 (1992). Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 200 KDa |
| Recombinant expression | Organism:  |
| Sequence | InterPro: Pyruvate dehydrogenase E1 component |
-Macromolecule #3: E3
| Macromolecule | Name: E3 / type: protein_or_peptide / ID: 3 / Name.synonym: dihydrolipoamide dehydrogenase Details: Up to 24 of these; See Wagenknecht, T. et al., JSB 109:70-77 (1992). Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 100 KDa |
| Recombinant expression | Organism:  |
| Sequence | InterPro: Dihydrolipoamide dehydrogenase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: 20 mM Potassium Phosphate |
| Grid | Details: R 1.5/1.3 Quantifoil |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: OTHER Details: Vitrification instrument: Vitrobot. Sample was at 22 C in air before plunging. Method: Blot for 3.5 seconds with an offset of -3 before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Min: 82 K / Max: 82 K / Average: 82 K |
| Alignment procedure | Legacy - Electron beam tilt params: 0 |
| Specialist optics | Energy filter - Name: GIF 3000 / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Date | Aug 24, 2004 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Average electron dose: 110 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 36600 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 10.0 µm / Nominal defocus min: 10.0 µm / Nominal magnification: 27500 |
| Sample stage | Specimen holder: FEI Polara / Specimen holder model: GATAN HELIUM / Tilt series - Axis1 - Min angle: -63 ° / Tilt series - Axis1 - Max angle: 66 ° |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Dual-axis tilt series, with 44 sections on one axis, and 45 on the other. Average number of tilts used in the 3D reconstructions: 89. Average tomographic tilt angle increment: 3. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 55.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name:  IMOD IMODDetails: The individual, orthogonal tomograms were filtered at their first CTF zero and then merged. |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)