[English] 日本語
 Yorodumi
Yorodumi- EMDB-1120: Conservation of the capsid structure in tailed dsDNA bacteriophag... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1120 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. | |||||||||
 Map data Map data | This map is a 3D cryo-EM reconstruction of a fiberless, isometric variant of bacteriophage phi29. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Bacillus phage phi29 (virus) Bacillus phage phi29 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.9 Å | |||||||||
 Authors Authors | Morais MC / Choi KH / Koti JS / Chipman PR / Anderson DL / Rossmann MG | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2005 Journal: Mol Cell / Year: 2005Title: Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Authors: Marc C Morais / Kyung H Choi / Jaya S Koti / Paul R Chipman / Dwight L Anderson / Michael G Rossmann /  Abstract: Bacteriophage phi29 is one of the smallest and simplest known dsDNA phages, making it amenable to structural investigations. The three-dimensional structure of a fiberless, isometric variant has been ...Bacteriophage phi29 is one of the smallest and simplest known dsDNA phages, making it amenable to structural investigations. The three-dimensional structure of a fiberless, isometric variant has been determined to 7.9 A resolution by cryo-electron microscopy (cryo-EM), allowing the identification of alpha helices and beta sheets. Their arrangement indicates that the folds of the phi29 and bacteriophage HK97 capsid proteins are similar except for an additional immunoglobulin-like domain of the phi29 protein. An atomic model that incorporates these two domains fits well into the cryo-EM density of the T = 3, fiberless isometric phi29 particle, and cryo-EM structures of fibered isometric and fiberless prolate prohead phi29 particles at resolutions of 8.7 A and 12.7 A, respectively. Thus, phi29 joins the growing number of phages that utilize the HK97 capsid structure, suggesting that this protein fold may be as prevalent in capsids of dsDNA phages as the jelly roll fold is in eukaryotic viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1120.map.gz emd_1120.map.gz | 4.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1120-v30.xml emd-1120-v30.xml emd-1120.xml emd-1120.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  1120.gif 1120.gif | 21 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1120 http://ftp.pdbj.org/pub/emdb/structures/EMD-1120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1120 | HTTPS FTP |
-Validation report
| Summary document |  emd_1120_validation.pdf.gz emd_1120_validation.pdf.gz | 404.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1120_full_validation.pdf.gz emd_1120_full_validation.pdf.gz | 404.4 KB | Display | |
| Data in XML |  emd_1120_validation.xml.gz emd_1120_validation.xml.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1120 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1120 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1120 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1120 | HTTPS FTP |
-Related structure data
| Related structure data |  1yxnMC  1116C  1117C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1120.map.gz / Format: CCP4 / Size: 82.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1120.map.gz / Format: CCP4 / Size: 82.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map is a 3D cryo-EM reconstruction of a fiberless, isometric variant of bacteriophage phi29. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
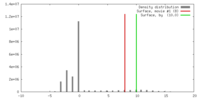
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : phi29 fiberless isometric particle
| Entire | Name: phi29 fiberless isometric particle |
|---|---|
| Components |
|
-Supramolecule #1000: phi29 fiberless isometric particle
| Supramolecule | Name: phi29 fiberless isometric particle / type: sample / ID: 1000 / Oligomeric state: particle forms an T3 icosahedron / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 8.97 MDa |
-Supramolecule #1: Bacillus phage phi29
| Supramolecule | Name: Bacillus phage phi29 / type: virus / ID: 1 / Name.synonym: phi29 / NCBI-ID: 10756 / Sci species name: Bacillus phage phi29 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes / Syn species name: phi29 |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Experimental: 8.97 MDa |
| Virus shell | Shell ID: 1 / Diameter: 425 Å / T number (triangulation number): 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Details: 25 mM Tris-HCl 5 mM MgCl2 50 mM NaCl 5 mM sodium azide |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Temperature | Average: 90.0 K |
| Alignment procedure | Legacy - Electron beam tilt params: 0 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 15 / Average electron dose: 20 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 38000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN / Tilt angle min: 0 / Tilt angle max: 0 |
- Image processing
Image processing
| CTF correction | Details: CTF correction included phase and amplitude correction for whole micrographs |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 7.9 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMPFT, POR, P3DR / Number images used: 5922 |
-Atomic model buiding 1
| Software | Name: SITUS and EMFIT |
|---|---|
| Details | Protocol: Rigid Body. The fitted molecule consists of two domains, an HK97-like domain and a bacterial immunoglobulin group 2-like domain (BIG2). The two domains were fitted separately. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT Target criteria: correlation of laplacian filtered data for SITUS, and for EMFIT, target maximizes positive density around an atom, minimizes negative density, and minimizes clashes with neighboring olecules |
| Output model |  PDB-1yxn: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)