[English] 日本語
 Yorodumi
Yorodumi- EMDB-10801: Folding of extension domain in duck hepatitis B virus capsids is ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10801 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Folding of extension domain in duck hepatitis B virus capsids is accelerated by FkpA | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Hepatitis B virus duck/DHBV-16 Hepatitis B virus duck/DHBV-16 | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Makbul C / Bottcher B | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Slowly folding surface extension in the prototypic avian hepatitis B virus capsid governs stability. Authors: Cihan Makbul / Michael Nassal / Bettina Böttcher /  Abstract: Hepatitis B virus (HBV) is an important but difficult to study human pathogen. Most basics of the hepadnaviral life-cycle were unraveled using duck HBV (DHBV) as a model although DHBV has a capsid ...Hepatitis B virus (HBV) is an important but difficult to study human pathogen. Most basics of the hepadnaviral life-cycle were unraveled using duck HBV (DHBV) as a model although DHBV has a capsid protein (CP) comprising ~260 rather than ~180 amino acids. Here we present high-resolution structures of several DHBV capsid-like particles (CLPs) determined by electron cryo-microscopy. As for HBV, DHBV CLPs consist of a dimeric α-helical frame-work with protruding spikes at the dimer interface. A fundamental new feature is a ~ 45 amino acid proline-rich extension in each monomer replacing the tip of the spikes in HBV CP. In vitro, folding of the extension takes months, implying a catalyzed process in vivo. DHBc variants lacking a folding-proficient extension produced regular CLPs in bacteria but failed to form stable nucleocapsids in hepatoma cells. We propose that the extension domain acts as a conformational switch with differential response options during viral infection. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10801.map.gz emd_10801.map.gz | 227.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10801-v30.xml emd-10801-v30.xml emd-10801.xml emd-10801.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10801_fsc.xml emd_10801_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10801.png emd_10801.png | 318.1 KB | ||
| Masks |  emd_10801_msk_1.map emd_10801_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Others |  emd_10801_half_map_1.map.gz emd_10801_half_map_1.map.gz emd_10801_half_map_2.map.gz emd_10801_half_map_2.map.gz | 202.6 MB 202.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10801 http://ftp.pdbj.org/pub/emdb/structures/EMD-10801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10801 | HTTPS FTP |
-Validation report
| Summary document |  emd_10801_validation.pdf.gz emd_10801_validation.pdf.gz | 415.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10801_full_validation.pdf.gz emd_10801_full_validation.pdf.gz | 414.3 KB | Display | |
| Data in XML |  emd_10801_validation.xml.gz emd_10801_validation.xml.gz | 20.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10801 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10801 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10801 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10801 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10801.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10801.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0635 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
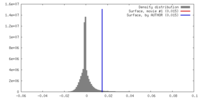
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10801_msk_1.map emd_10801_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
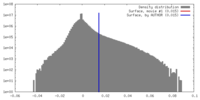
| Density Histograms |
-Half map: #2
| File | emd_10801_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10801_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hepatitis B virus duck/DHBV-16
| Entire | Name:  Hepatitis B virus duck/DHBV-16 Hepatitis B virus duck/DHBV-16 |
|---|---|
| Components |
|
-Supramolecule #1: Hepatitis B virus duck/DHBV-16
| Supramolecule | Name: Hepatitis B virus duck/DHBV-16 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: TheDuck Hepatitis B core protein was co-expressed with FKPB-type peptidyl-prolyl cis-trans isomerase FkpA to enhance folding of the proline rich extension domain. Co-expression with FkpA was ...Details: TheDuck Hepatitis B core protein was co-expressed with FKPB-type peptidyl-prolyl cis-trans isomerase FkpA to enhance folding of the proline rich extension domain. Co-expression with FkpA was conducted analogously to HBc-SRPK1 co-expression (Heger-Stevic et al. PlosPath 2018), using anhydrotetracyclin for Tet promoter induction. NCBI-ID: 489543 / Sci species name: Hepatitis B virus duck/DHBV-16 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Host system | Organism:  |
| Molecular weight | Theoretical: 7.2 MDa |
| Virus shell | Shell ID: 1 / Name: duck Hepatitis B Virus capsid / Diameter: 370.0 Å / T number (triangulation number): 4 |
-Macromolecule #1: Duck hepatitis B virus capsid protein co-expressed with peptidyl-...
| Macromolecule | Name: Duck hepatitis B virus capsid protein co-expressed with peptidyl-prolyl cis-trans isomerase FkpA type: protein_or_peptide / ID: 1 Details: Duck Hepatitis B capsid protein was co-expressed with FkpA to enhance folding. FkpA is not part of the assembly Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Hepatitis B virus duck/DHBV-16 Hepatitis B virus duck/DHBV-16 |
| Recombinant expression | Organism:  |
| Sequence | String: MDINASRALA NVYDLPDDFF PKIDDLVRDA KDALEPYWKS DSIKKHVLIA THFVDLIEDF WQTTQGMHEI AESLRAVIPP TTTPVPPGYL IQHEEAEEIP LGDLFKHQEE RIVSFQPDYP ITARIHAHLK AYAKINEESL DRARRLLWWH YNCLLWGEAQ VTNYISRLRT ...String: MDINASRALA NVYDLPDDFF PKIDDLVRDA KDALEPYWKS DSIKKHVLIA THFVDLIEDF WQTTQGMHEI AESLRAVIPP TTTPVPPGYL IQHEEAEEIP LGDLFKHQEE RIVSFQPDYP ITARIHAHLK AYAKINEESL DRARRLLWWH YNCLLWGEAQ VTNYISRLRT WLSTPEKYRG RDAPTIEAIT RPIQVAQGGR KTTTGTRKPR GLEPRRRKVK TTVVYGRRRS KSRERRAPTP QRAGSPLPRS SSSHHRSPSP RK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3. mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: For the vitrification, grids (400 mesh copper grids (type R 1.2/1.3. Quantifoil Micro Tools, Jena/Germany) were rendered hydrophilic by glow discharging in air at a pressure of 29 Pa for 2 ...Details: For the vitrification, grids (400 mesh copper grids (type R 1.2/1.3. Quantifoil Micro Tools, Jena/Germany) were rendered hydrophilic by glow discharging in air at a pressure of 29 Pa for 2 minutes at medium power with a Plasma Cleaner (model PDC-002. Harrick Plasma Ithaca, NY/USA). Then, 3.5 ul of DHBc solution was pipetted onto the grids and they were plunge frozen in liquid ethane with a Vitrobot mark IV (FEI-Thermo Fisher Scientific). The settings for the Vitrobot were 3s blot time, 45 s wait time, blot force 0 at a temperature of 4 C and 100 % humidity. | ||||||||||||||||||
| Details | DHBc was co-expressed with FkpA to enhance folding of the extension domain. The sample is prepared for cryo-EM after 2 weeks of storage. The extension domain is fully folded, which is in contrast to an largely unfolded extension domain in DHBc without co-expression of FkpA after 2 weeks of storage. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 4339 / Average exposure time: 2.4 sec. / Average electron dose: 40.0 e/Å2 Details: movie mode, 3 images per hole, 40 fractions per movie |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 1.9000000000000001 µm / Calibrated defocus min: 0.6 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)