[English] 日本語
 Yorodumi
Yorodumi- EMDB-10104: Structure of Azospirillum brasilense Glutamate Synthase in a4b4 o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10104 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Azospirillum brasilense Glutamate Synthase in a4b4 oligomeric state. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glutamate synthesys / complex / oligomeric assemblies / FLAVOPROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglutamate synthase (NADPH) / glutamate synthase (NADPH) activity / L-glutamate biosynthetic process / ammonia assimilation cycle / 3 iron, 4 sulfur cluster binding / 4 iron, 4 sulfur cluster binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Azospirillum brasilense (bacteria) Azospirillum brasilense (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Swuec P | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2019 Journal: J Mol Biol / Year: 2019Title: Cryo-EM Structures of Azospirillum brasilense Glutamate Synthase in Its Oligomeric Assemblies. Authors: Paolo Swuec / Antonio Chaves-Sanjuan / Carlo Camilloni / Maria Antonietta Vanoni / Martino Bolognesi /  Abstract: Bacterial NADPH-dependent glutamate synthase (GltS) is a complex iron-sulfur flavoprotein that catalyzes the reductive synthesis of two L-Glu molecules from L-Gln and 2-oxo-glutarate. GltS functional ...Bacterial NADPH-dependent glutamate synthase (GltS) is a complex iron-sulfur flavoprotein that catalyzes the reductive synthesis of two L-Glu molecules from L-Gln and 2-oxo-glutarate. GltS functional unit hosts an α-subunit (αGltS) and a β-subunit (βGltS) that assemble in different αβ oligomers in solution. Here, we present the cryo-electron microscopy structures of Azospirillum brasilense GltS in four different oligomeric states (αβ, αβ, αβ and αβ, in the 3.5- to 4.1-Å resolution range). Our study provides a comprehensive GltS model that details the inter-protomeric assemblies and allows unequivocal location of the FAD cofactor and of two electron transfer [4Fe-4S] clusters within βGltS. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10104.map.gz emd_10104.map.gz | 104.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10104-v30.xml emd-10104-v30.xml emd-10104.xml emd-10104.xml | 26.7 KB 26.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10104_fsc.xml emd_10104_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_10104.png emd_10104.png | 167.6 KB | ||
| Masks |  emd_10104_msk_1.map emd_10104_msk_1.map | 111.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10104.cif.gz emd-10104.cif.gz | 7.7 KB | ||
| Others |  emd_10104_additional.map.gz emd_10104_additional.map.gz emd_10104_half_map_1.map.gz emd_10104_half_map_1.map.gz emd_10104_half_map_2.map.gz emd_10104_half_map_2.map.gz | 87.5 MB 87.7 MB 87.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10104 http://ftp.pdbj.org/pub/emdb/structures/EMD-10104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10104 | HTTPS FTP |
-Validation report
| Summary document |  emd_10104_validation.pdf.gz emd_10104_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10104_full_validation.pdf.gz emd_10104_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_10104_validation.xml.gz emd_10104_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_10104_validation.cif.gz emd_10104_validation.cif.gz | 23.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10104 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10104 | HTTPS FTP |
-Related structure data
| Related structure data |  6s6sMC  6s6tC  6s6uC  6s6xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10104.map.gz / Format: CCP4 / Size: 111.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10104.map.gz / Format: CCP4 / Size: 111.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.426 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10104_msk_1.map emd_10104_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
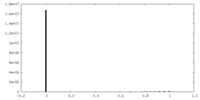
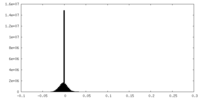
| Density Histograms |
-Additional map: #1
| File | emd_10104_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
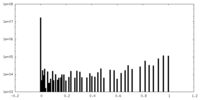
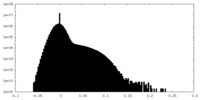
| Density Histograms |
-Half map: #1
| File | emd_10104_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
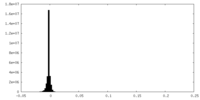
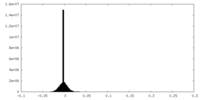
| Density Histograms |
-Half map: #2
| File | emd_10104_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
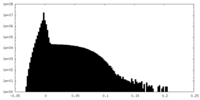
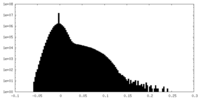
| Density Histograms |
- Sample components
Sample components
-Entire : Glutamate Synthase complex in a4b4 oligomeric state
| Entire | Name: Glutamate Synthase complex in a4b4 oligomeric state |
|---|---|
| Components |
|
-Supramolecule #1: Glutamate Synthase complex in a4b4 oligomeric state
| Supramolecule | Name: Glutamate Synthase complex in a4b4 oligomeric state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Azospirillum brasilense (bacteria) Azospirillum brasilense (bacteria) |
-Macromolecule #1: Glutamate synthase [NADPH] large chain
| Macromolecule | Name: Glutamate synthase [NADPH] large chain / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: glutamate synthase (NADPH) |
|---|---|
| Source (natural) | Organism:  Azospirillum brasilense (bacteria) Azospirillum brasilense (bacteria) |
| Molecular weight | Theoretical: 166.224734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTELNQGEQ FVADFRANAA ALTTANAYNP EDEHDACGVG FIAAIDGKPR RSVVEKGIEA LKAVWHRGAV DADGKTGDGA GIHVAVPQK FFKDHVKVIG HRAPDNKLAV GQVFLPRISL DAQEACRCIV ETEILAFGYY IYGWRQVPIN VDIIGEKANA T RPEIEQII ...String: MTTELNQGEQ FVADFRANAA ALTTANAYNP EDEHDACGVG FIAAIDGKPR RSVVEKGIEA LKAVWHRGAV DADGKTGDGA GIHVAVPQK FFKDHVKVIG HRAPDNKLAV GQVFLPRISL DAQEACRCIV ETEILAFGYY IYGWRQVPIN VDIIGEKANA T RPEIEQII VGNNKGVSDE QFELDLYIIR RRIEKAVKGE QINDFYICSL SARSIIYKGM FLAEQLTTFY PDLLDERFES DF AIYHQRY STNTFPTWPL AQPFRMLAHN GEINTVKGNV NWMKAHETRM EHPAFGTHMQ DLKPVIGVGL SDSGSLDTVF EVM VRAGRT APMVKMMLVP QALTSSQTTP DNHKALIQYC NSVMEPWDGP AALAMTDGRW VVGGMDRNGL RPMRYTITTD GLII GGSET GMVKIDETQV IEKGRLGPGE MIAVDLQSGK LYRDRELKDH LATLKPWDKW VQNTTHLDEL VKTASLKGEP SDMDK AELR RRQQAFGLTM EDMELILHPM VEDGKEAIGS MGDDSPIAVL SDKYRGLHHF FRQNFSQVTN PPIDSLRERR VMSLKT RLG NLGNILDEDE TQTRLLQLES PVLTTAEFRA MRDYMGDTAA EIDATFPVDG GPEALRDALR RIRQETEDAV RGGATHV IL TDEAMGPARA AIPAILATGA VHTHLIRSNL RTFTSLNVRT AEGLDTHYFA VLIGVGATTV NAYLAQEAIA ERHRRGLF G SMPLEKGMAN YKKAIDDGLL KIMSKMGISV ISSYRGGGNF EAIGLSRALV AEHFPAMVSR ISGIGLNGIQ KKVLEQHAT AYNEEVVALP VGGFYRFRKS GDRHGWEGGV IHTLQQAVTN DSYTTFKKYS EQVNKRPPMQ LRDLLELRST KAPVPVDEVE SITAIRKRF ITPGMSMGAL SPEAHGTLNV AMNRIGAKSD SGEGGEDPAR FRPDKNGDNW NSAIKQVASG RFGVTAEYLN Q CRELEIKV AQGAKPGEGG QLPGFKVTEM IARLRHSTPG VMLISPPPHH DIYSIEDLAQ LIYDLKQINP DAKVTVKLVS RS GIGTIAA GVAKANADII LISGNSGGTG ASPQTSIKFA GLPWEMGLSE VHQVLTLNRL RHRVRLRTDG GLKTGRDIVI AAM LGAEEF GIGTASLIAM GCIMVRQCHS NTCPVGVCVQ DDKLRQKFVG TPEKVVNLFT FLAEEVREIL AGLGFRSLNE VIGR TDLLH QVSRGAEHLD DLDLNPRLAQ VDPGENARYC TLQGRNEVPD TLDARIVADA RPLFEEGEKM QLAYNARNTQ RAIGT RLSS MVTRKFGMFG LQPGHITIRL RGTAGQSLGA FAVQGIKLEV MGDANDYVGK GLSGGTIVVR PTTSSPLETN KNTIIG NTV LYGATAGKLF AAGQAGERFA VRNSGATVVV EGCGSNGCEY MTGGTAVILG RVGDNFAAGM TGGMAYVYDL DDSLPLY IN DESVIFQRIE VGHYESQLKH LIEEHVTETQ SRFAAEILND WAREVTKFWQ VVPKEMLNRL EVPVHLPKAI SAE UniProtKB: Glutamate synthase [NADPH] large chain |
-Macromolecule #2: Glutamate synthase [NADPH] small chain
| Macromolecule | Name: Glutamate synthase [NADPH] small chain / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: glutamate synthase (NADPH) |
|---|---|
| Source (natural) | Organism:  Azospirillum brasilense (bacteria) Azospirillum brasilense (bacteria) |
| Molecular weight | Theoretical: 52.425109 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANQRMLGFV HTAQRMPDKR PAAERRQDFA EIYARFSDER ANEQANRCSQ CGVPFCQVHC PVSNNIPDWL KLTSEGRLEE AYEVSQATN NFPEICGRIC PQDRLCEGNC VIEQSTHGAV TIGSVEKYIN DTAWDQGWVK PRTPSRELGL SVGVIGAGPA G LAAAEELR ...String: MANQRMLGFV HTAQRMPDKR PAAERRQDFA EIYARFSDER ANEQANRCSQ CGVPFCQVHC PVSNNIPDWL KLTSEGRLEE AYEVSQATN NFPEICGRIC PQDRLCEGNC VIEQSTHGAV TIGSVEKYIN DTAWDQGWVK PRTPSRELGL SVGVIGAGPA G LAAAEELR AKGYEVHVYD RYDRMGGLLV YGIPGFKLEK SVVERRVKLL ADAGVIYHPN FEVGRDASLP ELRRKHVAVL VA TGVYKAR DIKAPGSGLG NIVAALDYLT TSNKVSLGDT VEAYENGSLN AAGKHVVVLG GGDTAMDCVR TAIRQGATSV KCL YRRDRK NMPGSQREVA HAEEEGVEFI WQAAPEGFTG DTVVTGVRAV RIHLGVADAT GRQTPQVIEG SEFTVQADLV IKAL GFEPE DLPNAFDEPE LKVTRWGTLL VDHRTKMTNM DGVFAAGDIV RGASLVVWAI RDGRDAAEGI HAYAKAKAEA PVAVA AE UniProtKB: Glutamate synthase [NADPH] small chain |
-Macromolecule #3: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 4 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Macromolecule #4: FE3-S4 CLUSTER
| Macromolecule | Name: FE3-S4 CLUSTER / type: ligand / ID: 4 / Number of copies: 4 / Formula: F3S |
|---|---|
| Molecular weight | Theoretical: 295.795 Da |
| Chemical component information |  ChemComp-F3S: |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 8 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #6: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 6 / Number of copies: 4 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)