[English] 日本語
 Yorodumi
Yorodumi- EMDB-0598: Structure of a drug-like molecule stalled ribosome nascent chain ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0598 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a drug-like molecule stalled ribosome nascent chain complex (PCSK9_RNC) in non-rotated state with PP tRNA | |||||||||
 Map data Map data | PF846 stalled human ribosome nascent chain complex (PCSK9-RNC) in non-rotated state with PP tRNA. Map is sharpened. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationlow-density lipoprotein particle receptor catabolic process / negative regulation of receptor-mediated endocytosis involved in cholesterol transport / extrinsic component of external side of plasma membrane / negative regulation of sodium ion import across plasma membrane / PCSK9-LDLR complex / PCSK9-AnxA2 complex / negative regulation of receptor recycling / apolipoprotein receptor binding / very-low-density lipoprotein particle binding / positive regulation of low-density lipoprotein particle receptor catabolic process ...low-density lipoprotein particle receptor catabolic process / negative regulation of receptor-mediated endocytosis involved in cholesterol transport / extrinsic component of external side of plasma membrane / negative regulation of sodium ion import across plasma membrane / PCSK9-LDLR complex / PCSK9-AnxA2 complex / negative regulation of receptor recycling / apolipoprotein receptor binding / very-low-density lipoprotein particle binding / positive regulation of low-density lipoprotein particle receptor catabolic process / low-density lipoprotein particle binding / LDL clearance / lipoprotein metabolic process / very-low-density lipoprotein particle receptor binding / signaling receptor inhibitor activity / embryonic brain development / negative regulation of receptor internalization / translation at presynapse / COPII-coated ER to Golgi transport vesicle / exit from mitosis / optic nerve development / sodium channel inhibitor activity / response to insecticide / eukaryotic 80S initiation complex / endolysosome membrane / regulation of translation involved in cellular response to UV / negative regulation of protein neddylation / negative regulation of formation of translation preinitiation complex / axial mesoderm development / regulation of G1 to G0 transition / retinal ganglion cell axon guidance / negative regulation of endoplasmic reticulum unfolded protein response / ribosomal protein import into nucleus / oxidized pyrimidine DNA binding / response to TNF agonist / positive regulation of base-excision repair / positive regulation of respiratory burst involved in inflammatory response / negative regulation of low-density lipoprotein particle clearance / protein-DNA complex disassembly / positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / positive regulation of gastrulation / positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage / protein tyrosine kinase inhibitor activity / positive regulation of endodeoxyribonuclease activity / 90S preribosome assembly / nucleolus organization / IRE1-RACK1-PP2A complex / positive regulation of Golgi to plasma membrane protein transport / TNFR1-mediated ceramide production / lysosomal transport / alpha-beta T cell differentiation / GAIT complex / negative regulation of RNA splicing / negative regulation of DNA repair / positive regulation of DNA damage response, signal transduction by p53 class mediator / TORC2 complex binding / triglyceride metabolic process / G1 to G0 transition / supercoiled DNA binding / NF-kappaB complex / low-density lipoprotein particle receptor binding / cysteine-type endopeptidase activator activity involved in apoptotic process / neural crest cell differentiation / oxidized purine DNA binding / positive regulation of ubiquitin-protein transferase activity / middle ear morphogenesis / negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide / negative regulation of bicellular tight junction assembly / regulation of establishment of cell polarity / ubiquitin-like protein conjugating enzyme binding / rRNA modification in the nucleus and cytosol / erythrocyte homeostasis / negative regulation of phagocytosis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / negative regulation of ubiquitin protein ligase activity / protein kinase A binding / homeostatic process / laminin receptor activity / ion channel inhibitor activity / Ribosomal scanning and start codon recognition / pigmentation / Translation initiation complex formation / positive regulation of mitochondrial depolarization / macrophage chemotaxis / protein autoprocessing / lung morphogenesis / cellular response to actinomycin D / fibroblast growth factor binding / positive regulation of T cell receptor signaling pathway / negative regulation of Wnt signaling pathway / positive regulation of natural killer cell proliferation / monocyte chemotaxis / negative regulation of translational frameshifting / TOR signaling / BH3 domain binding / Protein hydroxylation / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / positive regulation of receptor internalization / positive regulation of activated T cell proliferation Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
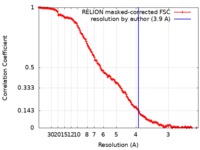
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Li W / Cate JHD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Structural basis for selective stalling of human ribosome nascent chain complexes by a drug-like molecule. Authors: Wenfei Li / Fred R Ward / Kim F McClure / Stacey Tsai-Lan Chang / Elizabeth Montabana / Spiros Liras / Robert G Dullea / Jamie H D Cate /  Abstract: The drug-like molecule PF-06446846 (PF846) binds the human ribosome and selectively blocks the translation of a small number of proteins by an unknown mechanism. In structures of PF846-stalled human ...The drug-like molecule PF-06446846 (PF846) binds the human ribosome and selectively blocks the translation of a small number of proteins by an unknown mechanism. In structures of PF846-stalled human ribosome nascent chain complexes, PF846 binds in the ribosome exit tunnel in a eukaryotic-specific pocket formed by 28S ribosomal RNA, and alters the path of the nascent polypeptide chain. PF846 arrests the translating ribosome in the rotated state of translocation, in which the peptidyl-transfer RNA 3'-CCA end is improperly docked in the peptidyl transferase center. Selections of messenger RNAs from mRNA libraries using translation extracts reveal that PF846 can stall translation elongation, arrest termination or even enhance translation, depending on nascent chain sequence context. These results illuminate how a small molecule selectively targets translation by the human ribosome, and provides a foundation for developing small molecules that modulate the production of proteins of therapeutic interest. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0598.map.gz emd_0598.map.gz | 229 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0598-v30.xml emd-0598-v30.xml emd-0598.xml emd-0598.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0598_fsc.xml emd_0598_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0598.png emd_0598.png | 163.5 KB | ||
| Others |  emd_0598_additional.map.gz emd_0598_additional.map.gz emd_0598_half_map_1.map.gz emd_0598_half_map_1.map.gz emd_0598_half_map_2.map.gz emd_0598_half_map_2.map.gz | 193.1 MB 193.8 MB 193.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0598 http://ftp.pdbj.org/pub/emdb/structures/EMD-0598 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0598 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0598 | HTTPS FTP |
-Related structure data
| Related structure data |  6olzMC  0526C  0596C  0597C  0599C  0600C  0601C  6oleC  6olfC  6olgC  6oliC  6om0C  6om7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0598.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0598.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PF846 stalled human ribosome nascent chain complex (PCSK9-RNC) in non-rotated state with PP tRNA. Map is sharpened. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.22 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
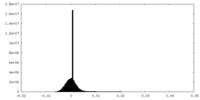
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: PF846 stalled human ribosome nascent chain complex (PCSK9-RNC)...
| File | emd_0598_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PF846 stalled human ribosome nascent chain complex (PCSK9-RNC) in non-rotated state with PP tRNA. Map is not sharpened. | ||||||||||||
| Projections & Slices |
| ||||||||||||
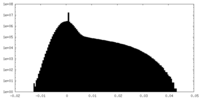
| Density Histograms |
-Half map: Half map of PF846 stalled human ribosome nascent...
| File | emd_0598_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of PF846 stalled human ribosome nascent chain complex (PCSK9-RNC) in non-rotated state with PP tRNA. | ||||||||||||
| Projections & Slices |
| ||||||||||||
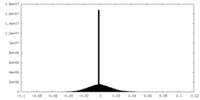
| Density Histograms |
-Half map: Half map of PF846 stalled human ribosome nascent...
| File | emd_0598_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of PF846 stalled human ribosome nascent chain complex (PCSK9-RNC) in non-rotated state with PP tRNA. | ||||||||||||
| Projections & Slices |
| ||||||||||||
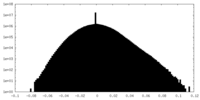
| Density Histograms |
- Sample components
Sample components
-Entire : Drug-like molecule stalled human ribosome nascent chain complex (...
| Entire | Name: Drug-like molecule stalled human ribosome nascent chain complex (PCSK9-RNC) in non-rotated state with PP tRNA |
|---|---|
| Components |
|
-Supramolecule #1: Drug-like molecule stalled human ribosome nascent chain complex (...
| Supramolecule | Name: Drug-like molecule stalled human ribosome nascent chain complex (PCSK9-RNC) in non-rotated state with PP tRNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HeLa Homo sapiens (human) / Recombinant cell: HeLa |
| Molecular weight | Theoretical: 4.3 MDa |
-Macromolecule #1: human ribosome with nascent chain
| Macromolecule | Name: human ribosome with nascent chain / type: rna / ID: 1 Details: Nascent chain sequence including CDH1 domain V and PCSK9 1-35 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SDVNDNAPIP EPRTIFFCER NPKPQVINII DADLPPNTSP FTAELTHGAS ANWTIQYNDP TQESIILKPK MALEVGDYKI NLKLMDNQNK DQVTTLEVSV CDCEGAAGVC RKAQPVEAGL QIPAILGILG MGTVSSRRS WWPLPLLLLL LLLLGPAGAR AQEDED |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Overall B value: 80 |
|---|---|
| Output model |  PDB-6olz: |
 Movie
Movie Controller
Controller















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)