[English] 日本語
 Yorodumi
Yorodumi- EMDB-0433: AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targe... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0433 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody ACS202 fragment antigen binding | ||||||||||||||||||
 Map data Map data | sharpened map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | HIV-1 Env / SOSIP / trimer / broadly neutralizing antibody / fusion peptide / VIRAL PROTEIN / IMMUNE SYSTEM | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationclathrin-dependent endocytosis of virus by host cell / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity Similarity search - Function | ||||||||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
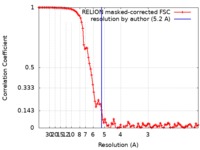
| Method | single particle reconstruction / cryo EM / Resolution: 5.2 Å | ||||||||||||||||||
 Authors Authors | Ozorowski G / de Val N / Cottrell CA / Copps J / Ward AB | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2019 Journal: Cell Host Microbe / Year: 2019Title: Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies. Authors: Meng Yuan / Christopher A Cottrell / Gabriel Ozorowski / Marit J van Gils / Sonu Kumar / Nicholas C Wu / Anita Sarkar / Jonathan L Torres / Natalia de Val / Jeffrey Copps / John P Moore / ...Authors: Meng Yuan / Christopher A Cottrell / Gabriel Ozorowski / Marit J van Gils / Sonu Kumar / Nicholas C Wu / Anita Sarkar / Jonathan L Torres / Natalia de Val / Jeffrey Copps / John P Moore / Rogier W Sanders / Andrew B Ward / Ian A Wilson /   Abstract: The fusion peptide (FP) of HIV-1 envelope glycoprotein (Env) is essential for mediating viral entry. Detection of broadly neutralizing antibodies (bnAbs) that interact with the FP has revealed it as ...The fusion peptide (FP) of HIV-1 envelope glycoprotein (Env) is essential for mediating viral entry. Detection of broadly neutralizing antibodies (bnAbs) that interact with the FP has revealed it as a site of vulnerability. We delineate X-ray and cryo-electron microscopy (cryo-EM) structures of bnAb ACS202, from an HIV-infected elite neutralizer, with an FP and with a soluble Env trimer (AMC011 SOSIP.v4.2) derived from the same patient. We show that ACS202 CDRH3 forms a "β strand" interaction with the exposed hydrophobic FP and recognizes a continuous region of gp120, including a conserved N-linked glycan at N88. A cryo-EM structure of another previously identified bnAb VRC34.01 with AMC011 SOSIP.v4.2 shows that it also penetrates through glycans to target the FP. We further demonstrate that the FP can twist and present different conformations for recognition by bnAbs, which enables approach to Env from diverse angles. The variable recognition of FP by bnAbs thus provides insights for vaccine design. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0433.map.gz emd_0433.map.gz | 84.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0433-v30.xml emd-0433-v30.xml emd-0433.xml emd-0433.xml | 25.2 KB 25.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0433_fsc.xml emd_0433_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_0433.png emd_0433.png | 97.2 KB | ||
| Masks |  emd_0433_msk_1.map emd_0433_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0433.cif.gz emd-0433.cif.gz | 7.6 KB | ||
| Others |  emd_0433_half_map_1.map.gz emd_0433_half_map_1.map.gz emd_0433_half_map_2.map.gz emd_0433_half_map_2.map.gz | 70.9 MB 71 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0433 http://ftp.pdbj.org/pub/emdb/structures/EMD-0433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0433 | HTTPS FTP |
-Related structure data
| Related structure data |  6nc2MC  0434C  6nc3C  6ncpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0433.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0433.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
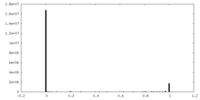
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0433_msk_1.map emd_0433_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
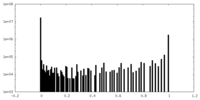
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_0433_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
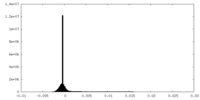
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_0433_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
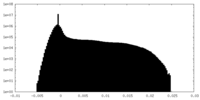
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of AMC011 v4.2 SOSIP trimer and ACS202 fragment antigen b...
| Entire | Name: Complex of AMC011 v4.2 SOSIP trimer and ACS202 fragment antigen binding |
|---|---|
| Components |
|
-Supramolecule #1: Complex of AMC011 v4.2 SOSIP trimer and ACS202 fragment antigen b...
| Supramolecule | Name: Complex of AMC011 v4.2 SOSIP trimer and ACS202 fragment antigen binding type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Supramolecule #2: HIV-1 Env AMC011 v4.2 SOSIP gp120
| Supramolecule | Name: HIV-1 Env AMC011 v4.2 SOSIP gp120 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #3: HIV-1 Env AMC011 v4.2 SOSIP gp41
| Supramolecule | Name: HIV-1 Env AMC011 v4.2 SOSIP gp41 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #4: VRC34.01
| Supramolecule | Name: VRC34.01 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: AMC011 v4.2 SOSIP gp120
| Macromolecule | Name: AMC011 v4.2 SOSIP gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 57.668938 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAEQLW VTVYYGVPVW KEATTTLFCA SDARAYDTEV RNVWATHACV PTDPNPQEV VLENVTENFN MWKNNMVEQM HEDIISLWDQ SLKPCVKLTP LCVTLNCTDL RNATNTNATN TTSSSRGTME G GEIKNCSF ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAEQLW VTVYYGVPVW KEATTTLFCA SDARAYDTEV RNVWATHACV PTDPNPQEV VLENVTENFN MWKNNMVEQM HEDIISLWDQ SLKPCVKLTP LCVTLNCTDL RNATNTNATN TTSSSRGTME G GEIKNCSF NITTSMRDKV QKEYALFYKL DVVPIKNDNT SYRLISCNTS VITQACPKVS FEPIPIHYCA PAGFAILKCN DK KFNGTGP CTNVSTVQCT HGIRPVVSTQ LLLNGSLAEE EVVIRSANFT DNAKIIIVQL NKSVEINCTR PNNNTRKSIH IGP GRWFYT TGEIIGDIRQ AHCNISGTKW NDTLKQIVVK LKEQFGNKTI VFNHSSGGDP EIVMHSFNCG GEFFYCNSTQ LFNS TWNDG SNYTGTIVLP CRIKQIVNMW QEVGKAMYAP PIKGQIRCSS NITGLILIRD GGKNRSENTE IFRPGGGDMR DNWRS ELYK YKVVKIEPLG IAPTKCKRRV VQRRRRRR |
-Macromolecule #2: AMC011 v4.2 SOSIP gp41
| Macromolecule | Name: AMC011 v4.2 SOSIP gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.300666 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RQLLSGIVQQ QNNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLKDQQ LLGIWGCSG KLICCTAVPW NTSWSNKSYN QIWNNMTWME WEREIDNYTS LIYTLIEDSQ NQQEKNEQEL LELD |
-Macromolecule #3: Monoclonal antibody ACS202 fragment antigen binding heavy chain
| Macromolecule | Name: Monoclonal antibody ACS202 fragment antigen binding heavy chain type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.392023 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MELGLRWVFL VAILEVHSQV QLVESGGGVV QPGGSLRLSC AASGFAFKDF GMHWVRQAPG KGLEWVAVIG GGHGQHQSYS ESVKGRFAI TRDNEKNKLY LHMDRLRTED TAVYYCAKDR LGRPWNIGGR LVYYYYGMDV WGQGTTVTVS SASTKGPSVF P LAPSSKST ...String: MELGLRWVFL VAILEVHSQV QLVESGGGVV QPGGSLRLSC AASGFAFKDF GMHWVRQAPG KGLEWVAVIG GGHGQHQSYS ESVKGRFAI TRDNEKNKLY LHMDRLRTED TAVYYCAKDR LGRPWNIGGR LVYYYYGMDV WGQGTTVTVS SASTKGPSVF P LAPSSKST SGGTAALGCL VKDYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PS NTKVDKK VEPKSCD |
-Macromolecule #4: Monoclonal antibody ACS202 fragment antigen binding kappa chain
| Macromolecule | Name: Monoclonal antibody ACS202 fragment antigen binding kappa chain type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.475658 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVHCA IRMTQSPSSL SASVGDRVTI TCQASQDIKK SLNWYRQKPG KAPELLIHDA SILQTGVPSA FTASGSGTH FSFVINKLQP EDVGTYFCQE YENLQFTFGP GTKVEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY P REAKVQWK ...String: MGWSCIILFL VATATGVHCA IRMTQSPSSL SASVGDRVTI TCQASQDIKK SLNWYRQKPG KAPELLIHDA SILQTGVPSA FTASGSGTH FSFVINKLQP EDVGTYFCQE YENLQFTFGP GTKVEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY P REAKVQWK VDNALQSGNS QESVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 30 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Detergent (DDM) added immediately prior to grid preparation | ||||||||||||
| Grid | Model: C-flat-2/2 4C / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER / Details: Gatan Solarus 950 Plasma system | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER | ||||||||||||
| Details | SEC purified sample after overnight incubation of molar excess Fab to trimer |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Number real images: 1641 / Average exposure time: 10.0 sec. / Average electron dose: 92.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6nc2: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)