[English] 日本語
 Yorodumi
Yorodumi- EMDB-0299: Bacteriophage phi6 nucleocapsid reconstructed with icosahedral sy... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0299 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry | |||||||||
 Map data Map data | Bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus / dsRNA / capsid | |||||||||
| Function / homology | : / Major inner capsid protein P1 / T=2 icosahedral viral capsid / viral inner capsid / viral nucleocapsid / RNA binding / identical protein binding / Major inner protein P1 Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas phage phi6 (virus) Pseudomonas phage phi6 (virus) | |||||||||
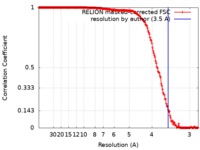
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Ilca SL / Huiskonen JT | |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus. Authors: Serban L Ilca / Xiaoyu Sun / Kamel El Omari / Abhay Kotecha / Felix de Haas / Frank DiMaio / Jonathan M Grimes / David I Stuart / Minna M Poranen / Juha T Huiskonen /     Abstract: Characterizing the genome of mature virions is pivotal to understanding the highly dynamic processes of virus assembly and infection. Owing to the different cellular fates of DNA and RNA, the life ...Characterizing the genome of mature virions is pivotal to understanding the highly dynamic processes of virus assembly and infection. Owing to the different cellular fates of DNA and RNA, the life cycles of double-stranded (ds)DNA and dsRNA viruses are dissimilar. In terms of nucleic acid packing, dsDNA viruses, which lack genome segmentation and intra-capsid transcriptional machinery, predominantly display single-spooled genome organizations. Because the release of dsRNA into the cytoplasm triggers host defence mechanisms, dsRNA viruses retain their genomes within a core particle that contains the enzymes required for RNA replication and transcription. The genomes of dsRNA viruses vary greatly in the degree of segmentation. In members of the Reoviridae family, genomes consist of 10-12 segments and exhibit a non-spooled arrangement mediated by RNA-dependent RNA polymerases. However, whether this arrangement is a general feature of dsRNA viruses remains unknown. Here, using cryo-electron microscopy to resolve the dsRNA genome structure of the tri-segmented bacteriophage ɸ6 of the Cystoviridae family, we show that dsRNA viruses can adopt a dsDNA-like single-spooled genome organization. We find that in this group of viruses, RNA-dependent RNA polymerases do not direct genome ordering, and the dsRNA can adopt multiple conformations. We build a model that encompasses 90% of the genome, and use this to quantify variation in the packing density and to characterize the different liquid crystalline geometries that are exhibited by the tightly compacted nucleic acid. Our results demonstrate that the canonical model for the packing of dsDNA can be extended to dsRNA viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0299.map.gz emd_0299.map.gz | 111.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0299-v30.xml emd-0299-v30.xml emd-0299.xml emd-0299.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0299_fsc.xml emd_0299_fsc.xml | 17.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0299.png emd_0299.png | 307.5 KB | ||
| Masks |  emd_0299_msk_1.map emd_0299_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0299.cif.gz emd-0299.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0299 http://ftp.pdbj.org/pub/emdb/structures/EMD-0299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0299 | HTTPS FTP |
-Validation report
| Summary document |  emd_0299_validation.pdf.gz emd_0299_validation.pdf.gz | 648.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0299_full_validation.pdf.gz emd_0299_full_validation.pdf.gz | 647.9 KB | Display | |
| Data in XML |  emd_0299_validation.xml.gz emd_0299_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_0299_validation.cif.gz emd_0299_validation.cif.gz | 22.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0299 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0299 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0299 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0299 | HTTPS FTP |
-Related structure data
| Related structure data |  6hy0MC  0294C  0295C  0296C  0300C  0302C  0304C  0305C  0306C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0299.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0299.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0299_msk_1.map emd_0299_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pseudomonas phage phi6
| Entire | Name:  Pseudomonas phage phi6 (virus) Pseudomonas phage phi6 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Pseudomonas phage phi6
| Supramolecule | Name: Pseudomonas phage phi6 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10879 / Sci species name: Pseudomonas phage phi6 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Pseudomonas syringae (bacteria) Pseudomonas syringae (bacteria) |
| Virus shell | Shell ID: 1 / Name: Outer Capsid Shell / T number (triangulation number): 13 |
| Virus shell | Shell ID: 2 / Name: Inner Capsid Shell / T number (triangulation number): 1 |
-Macromolecule #1: Major inner protein P1
| Macromolecule | Name: Major inner protein P1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phi6 (virus) Pseudomonas phage phi6 (virus) |
| Molecular weight | Theoretical: 85.080711 KDa |
| Sequence | String: MFNLKVKDLN GSARGLTQAF AIGELKNQLS VGALQLPLQF TRTFSASMTS ELLWEVGKGN IDPVMYARLF FQYAQAGGAL SVDELVNQF TEYHQSTACN PEIWRKLTAY ITGSSNRAIK ADAVGKVPPT AILEQLRTLA PSEHELFHHI TTDFVCHVLS P LGFILPDA ...String: MFNLKVKDLN GSARGLTQAF AIGELKNQLS VGALQLPLQF TRTFSASMTS ELLWEVGKGN IDPVMYARLF FQYAQAGGAL SVDELVNQF TEYHQSTACN PEIWRKLTAY ITGSSNRAIK ADAVGKVPPT AILEQLRTLA PSEHELFHHI TTDFVCHVLS P LGFILPDA AYVYRVGRTA TYPNFYALVD CVRASDLRRM LTALSSVDSK MLQATFKAKG ALAPALISQH LANAATTAFE RS RGNFDAN AVVSSVLTIL GRLWSPSTPK ELDPSARLRN TNGIDQLRSN LALFIAYQDM VKQRGRAEVI FSDEELSSTI IPW FIEAMS EVSPFKLRPI NETTSYIGQT SAIDHMGQPS HVVVYEDWQF AKEITAFTPV KLANNSNQRF LDVEPGISDR MSAT LAPIG NTFAVSAFVK NRTAVYEAVS QRGTVNSNGA EMTLGFPSVV ERDYALDRDP MVAIAALRTG IVDESLEARA SNDLK RSMF NYYAAVMHYA VAHNPEVVVS EHQGVAAEQG SLYLVWNVRT ELRIPVGYNA IEGGSIRTPE PLEAIAYNKP IQPSEV LQA KVLDLANHTT SIHIWPWHEA STEFAYEDAY SVTIRNKRYT AEVKEFELLG LGQRRERVRI LKPTVAHAII QMWYSWF VE DDRTLAAARR TSRDDAEKLA IDGRRMQNAV TLLRKIEMIG TTGIGASAVH LAQSRIVDQM AGRGLIDDSS DLHVGINR H RIRIWAGLAV LQMMGLLSRS EAEALTKVLG DSNALGMVVA TTDIDPSL UniProtKB: Major inner protein P1 |
-Macromolecule #2: Packaging Enzyme P4
| Macromolecule | Name: Packaging Enzyme P4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phi6 (virus) Pseudomonas phage phi6 (virus) |
| Molecular weight | Theoretical: 35.198426 KDa |
| Sequence | String: MPIVVTQAHI DRVGIAADLL DASPVSLQVL GRPTAINTVV IKTYIAAVME LASKQGGSLA GVDIRPSVLL KDTAIFTKPK AKSADVESD VDVLDTGIYS VPGLARKPVT HRWPSEGIYS GVTALMGATG SGKSITLNEK LRPDVLIRWG EVAEAYDELD T AVHISTLD ...String: MPIVVTQAHI DRVGIAADLL DASPVSLQVL GRPTAINTVV IKTYIAAVME LASKQGGSLA GVDIRPSVLL KDTAIFTKPK AKSADVESD VDVLDTGIYS VPGLARKPVT HRWPSEGIYS GVTALMGATG SGKSITLNEK LRPDVLIRWG EVAEAYDELD T AVHISTLD EMLIVCIGLG ALGFNVAVDS VRPLLFRLKG AASAGGIVAV FYSLLTDISN LFTQYDCSVV MVVNPMVDAE KI EYVFGQV MASTVGAILC ADGNVSRTMF RTNKGRIFNG AAPLAADTHM PSMDRPTSMK ALDHTSIASV APLERGSVDT DDR NSAPRR GANFSL |
-Macromolecule #3: Major Outer Capsid Protein P8
| Macromolecule | Name: Major Outer Capsid Protein P8 / type: protein_or_peptide / ID: 3 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phi6 (virus) Pseudomonas phage phi6 (virus) |
| Molecular weight | Theoretical: 16.018418 KDa |
| Sequence | String: MLLPVVARAA VPAIESAIAA TPGLVSRIAA AIGSKVSPSA ILAAVKSNPV VAGLTLAQIG STGYDAYQQL LENHPEVAEM LKDLSFKAD EIQPDFIGNL GQYREELELV EDAARFVGGM SNLIRLRQAL ELDIKYYGLK MQLNDMGYRS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)