[English] 日本語
 Yorodumi
Yorodumi- EMDB-0227: Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0227 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation | |||||||||
 Map data Map data | Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / serotonin receptor / pentameric ligand-gated channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationNeurotransmitter receptors and postsynaptic signal transmission / serotonin-gated cation-selective signaling pathway / serotonin-activated cation-selective channel complex / serotonin-gated monoatomic cation channel activity / serotonin receptor signaling pathway / serotonin binding / : / cleavage furrow / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / postsynaptic membrane / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Polovinkin L / Neumann E | |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Conformational transitions of the serotonin 5-HT receptor. Authors: Lucie Polovinkin / Ghérici Hassaine / Jonathan Perot / Emmanuelle Neumann / Anders A Jensen / Solène N Lefebvre / Pierre-Jean Corringer / Jacques Neyton / Christophe Chipot / Francois ...Authors: Lucie Polovinkin / Ghérici Hassaine / Jonathan Perot / Emmanuelle Neumann / Anders A Jensen / Solène N Lefebvre / Pierre-Jean Corringer / Jacques Neyton / Christophe Chipot / Francois Dehez / Guy Schoehn / Hugues Nury /    Abstract: The serotonin 5-HT receptor is a pentameric ligand-gated ion channel (pLGIC). It belongs to a large family of receptors that function as allosteric signal transducers across the plasma membrane; upon ...The serotonin 5-HT receptor is a pentameric ligand-gated ion channel (pLGIC). It belongs to a large family of receptors that function as allosteric signal transducers across the plasma membrane; upon binding of neurotransmitter molecules to extracellular sites, the receptors undergo complex conformational transitions that result in transient opening of a pore permeable to ions. 5-HT receptors are therapeutic targets for emesis and nausea, irritable bowel syndrome and depression. In spite of several reported pLGIC structures, no clear unifying view has emerged on the conformational transitions involved in channel gating. Here we report four cryo-electron microscopy structures of the full-length mouse 5-HT receptor in complex with the anti-emetic drug tropisetron, with serotonin, and with serotonin and a positive allosteric modulator, at resolutions ranging from 3.2 Å to 4.5 Å. The tropisetron-bound structure resembles those obtained with an inhibitory nanobody or without ligand. The other structures include an 'open' state and two ligand-bound states. We present computational insights into the dynamics of the structures, their pore hydration and free-energy profiles, and characterize movements at the gate level and cation accessibility in the pore. Together, these data deepen our understanding of the gating mechanism of pLGICs and capture ligand binding in unprecedented detail. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0227.map.gz emd_0227.map.gz | 5.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0227-v30.xml emd-0227-v30.xml emd-0227.xml emd-0227.xml | 10.6 KB 10.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0227.png emd_0227.png | 60 KB | ||
| Filedesc metadata |  emd-0227.cif.gz emd-0227.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0227 http://ftp.pdbj.org/pub/emdb/structures/EMD-0227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0227 | HTTPS FTP |
-Related structure data
| Related structure data |  6hiqMC  0225C  0226C  0228C  6hinC  6hioC  6hisC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0227.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0227.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.067 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 5-HT3 receptor, serotonin-bound
| Entire | Name: 5-HT3 receptor, serotonin-bound |
|---|---|
| Components |
|
-Supramolecule #1: 5-HT3 receptor, serotonin-bound
| Supramolecule | Name: 5-HT3 receptor, serotonin-bound / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 5-hydroxytryptamine receptor 3A
| Macromolecule | Name: 5-hydroxytryptamine receptor 3A / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.172305 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QPALLRLSDH LLANYKKGVR PVRDWRKPTT VSIDVIMYAI LNVDEKNQVL TTYIWYRQYW TDEFLQWTPE DFDNVTKLSI PTDSIWVPD ILINEFVDVG KSPNIPYVYV HHRGEVQNYK PLQLVTACSL DIYNFPFDVQ NCSLTFTSWL HTIQDINITL W RSPEEVRS ...String: QPALLRLSDH LLANYKKGVR PVRDWRKPTT VSIDVIMYAI LNVDEKNQVL TTYIWYRQYW TDEFLQWTPE DFDNVTKLSI PTDSIWVPD ILINEFVDVG KSPNIPYVYV HHRGEVQNYK PLQLVTACSL DIYNFPFDVQ NCSLTFTSWL HTIQDINITL W RSPEEVRS DKSIFINQGE WELLEVFPQF KEFSIDISNS YAEMKFYVII RRRPLFYAVS LLLPSIFLMV VDIVGFCLPP DS GERVSFK ITLLLGYSVF LIIVSDTLPA TAIGTPLIGV YFVVCMALLV ISLAETIFIV RLVHKQDLQR PVPDWLRHLV LDR IAWILC LGEQPMAHRP PATFQANKTD DCSGSDLLPA MGNHCSHVGG PQDLEKTPRG RGSPLPPPRE ASLAVRGLLQ ELSS IRHFL EKRDEMREVA RDWLRVGYVL DRLLFRIYLL AVLAYSITLV TLWSIWH UniProtKB: 5-hydroxytryptamine receptor 3A |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: SEROTONIN
| Macromolecule | Name: SEROTONIN / type: ligand / ID: 4 / Number of copies: 5 / Formula: SRO |
|---|---|
| Molecular weight | Theoretical: 176.215 Da |
| Chemical component information | 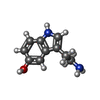 ChemComp-SRO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 62032 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















